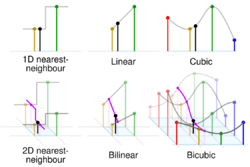
Bicubic interpolation
In mathematics, bicubic interpolation is an extension of cubic spline interpolation (a method of applying cubic interpolation to a data set) for interpolating data points on a two-dimensional regular grid. The interpolated surface (meaning the kernel shape, not the image) is smoother than corresponding surfaces obtained by bilinear interpolation or nearest-neighbor interpolation. Bicubic interpolation can be accomplished using either Lagrange polynomials, cubic splines, or cubic convolution algorithm.
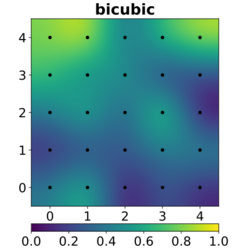
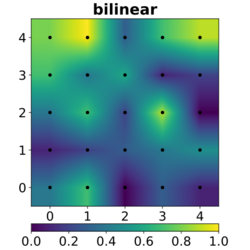
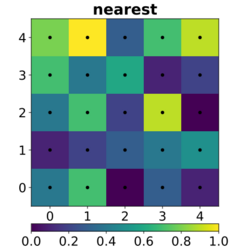
In image processing, bicubic interpolation is often chosen over bilinear or nearest-neighbor interpolation in image resampling, when speed is not an issue. In contrast to bilinear interpolation, which only takes 4 pixels (2×2) into account, bicubic interpolation considers 16 pixels (4×4). Images resampled with bicubic interpolation can have different interpolation artifacts, depending on the b and c values chosen.
Computation
Suppose the function values [math]\displaystyle{ f }[/math] and the derivatives [math]\displaystyle{ f_x }[/math], [math]\displaystyle{ f_y }[/math] and [math]\displaystyle{ f_{xy} }[/math] are known at the four corners [math]\displaystyle{ (0,0) }[/math], [math]\displaystyle{ (1,0) }[/math], [math]\displaystyle{ (0,1) }[/math], and [math]\displaystyle{ (1,1) }[/math] of the unit square. The interpolated surface can then be written as [math]\displaystyle{ p(x,y) = \sum\limits_{i=0}^3 \sum_{j=0}^3 a_{ij} x^i y^j. }[/math]
The interpolation problem consists of determining the 16 coefficients [math]\displaystyle{ a_{ij} }[/math]. Matching [math]\displaystyle{ p(x,y) }[/math] with the function values yields four equations:
- [math]\displaystyle{ f(0,0) = p(0,0) = a_{00}, }[/math]
- [math]\displaystyle{ f(1,0) = p(1,0) = a_{00} + a_{10} + a_{20} + a_{30}, }[/math]
- [math]\displaystyle{ f(0,1) = p(0,1) = a_{00} + a_{01} + a_{02} + a_{03}, }[/math]
- [math]\displaystyle{ f(1,1) = p(1,1) = \textstyle \sum\limits_{i=0}^3 \sum\limits_{j=0}^3 a_{ij}. }[/math]
Likewise, eight equations for the derivatives in the [math]\displaystyle{ x }[/math] and the [math]\displaystyle{ y }[/math] directions:
- [math]\displaystyle{ f_x(0,0) = p_x(0,0) = a_{10}, }[/math]
- [math]\displaystyle{ f_x(1,0) = p_x(1,0) = a_{10} + 2a_{20} + 3a_{30}, }[/math]
- [math]\displaystyle{ f_x(0,1) = p_x(0,1) = a_{10} + a_{11} + a_{12} + a_{13}, }[/math]
- [math]\displaystyle{ f_x(1,1) = p_x(1,1) = \textstyle \sum\limits_{i=1}^3 \sum\limits_{j=0}^3 a_{ij} i, }[/math]
- [math]\displaystyle{ f_y(0,0) = p_y(0,0) = a_{01}, }[/math]
- [math]\displaystyle{ f_y(1,0) = p_y(1,0) = a_{01} + a_{11} + a_{21} + a_{31}, }[/math]
- [math]\displaystyle{ f_y(0,1) = p_y(0,1) = a_{01} + 2a_{02} + 3a_{03}, }[/math]
- [math]\displaystyle{ f_y(1,1) = p_y(1,1) = \textstyle \sum\limits_{i=0}^3 \sum\limits_{j=1}^3 a_{ij} j. }[/math]
And four equations for the [math]\displaystyle{ xy }[/math] mixed partial derivative:
- [math]\displaystyle{ f_{xy}(0,0) = p_{xy}(0,0) = a_{11}, }[/math]
- [math]\displaystyle{ f_{xy}(1,0) = p_{xy}(1,0) = a_{11} + 2a_{21} + 3a_{31}, }[/math]
- [math]\displaystyle{ f_{xy}(0,1) = p_{xy}(0,1) = a_{11} + 2a_{12} + 3a_{13}, }[/math]
- [math]\displaystyle{ f_{xy}(1,1) = p_{xy}(1,1) = \textstyle \sum\limits_{i=1}^3 \sum\limits_{j=1}^3 a_{ij} i j. }[/math]
The expressions above have used the following identities: [math]\displaystyle{ p_x(x,y) = \textstyle \sum\limits_{i=1}^3 \sum\limits_{j=0}^3 a_{ij} i x^{i-1} y^j, }[/math] [math]\displaystyle{ p_y(x,y) = \textstyle \sum\limits_{i=0}^3 \sum\limits_{j=1}^3 a_{ij} x^i j y^{j-1}, }[/math] [math]\displaystyle{ p_{xy}(x,y) = \textstyle \sum\limits_{i=1}^3 \sum\limits_{j=1}^3 a_{ij} i x^{i-1} j y^{j-1}. }[/math]
This procedure yields a surface [math]\displaystyle{ p(x,y) }[/math] on the unit square [math]\displaystyle{ [0,1] \times [0,1] }[/math] that is continuous and has continuous derivatives. Bicubic interpolation on an arbitrarily sized regular grid can then be accomplished by patching together such bicubic surfaces, ensuring that the derivatives match on the boundaries.
Grouping the unknown parameters [math]\displaystyle{ a_{ij} }[/math] in a vector [math]\displaystyle{ \alpha=\left[\begin{smallmatrix}a_{00}&a_{10}&a_{20}&a_{30}&a_{01}&a_{11}&a_{21}&a_{31}&a_{02}&a_{12}&a_{22}&a_{32}&a_{03}&a_{13}&a_{23}&a_{33}\end{smallmatrix}\right]^T }[/math] and letting [math]\displaystyle{ x=\left[\begin{smallmatrix}f(0,0)&f(1,0)&f(0,1)&f(1,1)&f_x(0,0)&f_x(1,0)&f_x(0,1)&f_x(1,1)&f_y(0,0)&f_y(1,0)&f_y(0,1)&f_y(1,1)&f_{xy}(0,0)&f_{xy}(1,0)&f_{xy}(0,1)&f_{xy}(1,1)\end{smallmatrix}\right]^T, }[/math] the above system of equations can be reformulated into a matrix for the linear equation [math]\displaystyle{ A\alpha=x }[/math].
Inverting the matrix gives the more useful linear equation [math]\displaystyle{ A^{-1}x=\alpha }[/math], where [math]\displaystyle{ A^{-1}=\left[\begin{smallmatrix}\begin{array}{rrrrrrrrrrrrrrrr} 1 & 0 & 0 & 0 & 0 & 0 & 0 & 0 & 0 & 0 & 0 & 0 & 0 & 0 & 0 & 0 \\ 0 & 0 & 0 & 0 & 1 & 0 & 0 & 0 & 0 & 0 & 0 & 0 & 0 & 0 & 0 & 0 \\ -3 & 3 & 0 & 0 & -2 & -1 & 0 & 0 & 0 & 0 & 0 & 0 & 0 & 0 & 0 & 0 \\ 2 & -2 & 0 & 0 & 1 & 1 & 0 & 0 & 0 & 0 & 0 & 0 & 0 & 0 & 0 & 0 \\ 0 & 0 & 0 & 0 & 0 & 0 & 0 & 0 & 1 & 0 & 0 & 0 & 0 & 0 & 0 & 0 \\ 0 & 0 & 0 & 0 & 0 & 0 & 0 & 0 & 0 & 0 & 0 & 0 & 1 & 0 & 0 & 0 \\ 0 & 0 & 0 & 0 & 0 & 0 & 0 & 0 & -3 & 3 & 0 & 0 & -2 & -1 & 0 & 0 \\ 0 & 0 & 0 & 0 & 0 & 0 & 0 & 0 & 2 & -2 & 0 & 0 & 1 & 1 & 0 & 0 \\ -3 & 0 & 3 & 0 & 0 & 0 & 0 & 0 & -2 & 0 & -1 & 0 & 0 & 0 & 0 & 0 \\ 0 & 0 & 0 & 0 & -3 & 0 & 3 & 0 & 0 & 0 & 0 & 0 & -2 & 0 & -1 & 0 \\ 9 & -9 & -9 & 9 & 6 & 3 & -6 & -3 & 6 & -6 & 3 & -3 & 4 & 2 & 2 & 1 \\ -6 & 6 & 6 & -6 & -3 & -3 & 3 & 3 & -4 & 4 & -2 & 2 & -2 & -2 & -1 & -1 \\ 2 & 0 & -2 & 0 & 0 & 0 & 0 & 0 & 1 & 0 & 1 & 0 & 0 & 0 & 0 & 0 \\ 0 & 0 & 0 & 0 & 2 & 0 & -2 & 0 & 0 & 0 & 0 & 0 & 1 & 0 & 1 & 0 \\ -6 & 6 & 6 & -6 & -4 & -2 & 4 & 2 & -3 & 3 & -3 & 3 & -2 & -1 & -2 & -1 \\ 4 & -4 & -4 & 4 & 2 & 2 & -2 & -2 & 2 & -2 & 2 & -2 & 1 & 1 & 1 & 1 \end{array}\end{smallmatrix}\right], }[/math] which allows [math]\displaystyle{ \alpha }[/math] to be calculated quickly and easily.
There can be another concise matrix form for 16 coefficients: [math]\displaystyle{ \begin{bmatrix} f(0,0)&f(0,1)&f_y (0,0)&f_y (0,1)\\f(1,0)&f(1,1)&f_y (1,0)&f_y (1,1)\\f_x (0,0)&f_x (0,1)&f_{xy} (0,0)&f_{xy} (0,1)\\f_x (1,0)&f_x (1,1)&f_{xy} (1,0)&f_{xy} (1,1) \end{bmatrix} = \begin{bmatrix} 1&0&0&0\\1&1&1&1\\0&1&0&0\\0&1&2&3 \end{bmatrix} \begin{bmatrix} a_{00}&a_{01}&a_{02}&a_{03}\\a_{10}&a_{11}&a_{12}&a_{13}\\a_{20}&a_{21}&a_{22}&a_{23}\\a_{30}&a_{31}&a_{32}&a_{33} \end{bmatrix} \begin{bmatrix} 1&1&0&0\\0&1&1&1\\0&1&0&2\\0&1&0&3 \end{bmatrix}, }[/math] or [math]\displaystyle{ \begin{bmatrix} a_{00}&a_{01}&a_{02}&a_{03}\\a_{10}&a_{11}&a_{12}&a_{13}\\a_{20}&a_{21}&a_{22}&a_{23}\\a_{30}&a_{31}&a_{32}&a_{33} \end{bmatrix} = \begin{bmatrix} 1&0&0&0\\0&0&1&0\\-3&3&-2&-1\\2&-2&1&1 \end{bmatrix} \begin{bmatrix} f(0,0)&f(0,1)&f_y (0,0)&f_y (0,1)\\f(1,0)&f(1,1)&f_y (1,0)&f_y (1,1)\\f_x (0,0)&f_x (0,1)&f_{xy} (0,0)&f_{xy} (0,1)\\f_x (1,0)&f_x (1,1)&f_{xy} (1,0)&f_{xy} (1,1) \end{bmatrix} \begin{bmatrix} 1&0&-3&2\\0&0&3&-2\\0&1&-2&1\\0&0&-1&1 \end{bmatrix}, }[/math] where [math]\displaystyle{ p(x,y)=\begin{bmatrix}1 &x&x^2&x^3\end{bmatrix} \begin{bmatrix} a_{00}&a_{01}&a_{02}&a_{03}\\a_{10}&a_{11}&a_{12}&a_{13}\\a_{20}&a_{21}&a_{22}&a_{23}\\a_{30}&a_{31}&a_{32}&a_{33} \end{bmatrix} \begin{bmatrix}1\\y\\y^2\\y^3\end{bmatrix}. }[/math]
Extension to rectilinear grids
Often, applications call for bicubic interpolation using data on a rectilinear grid, rather than the unit square. In this case, the identities for [math]\displaystyle{ p_x, p_y, }[/math] and [math]\displaystyle{ p_{xy} }[/math] become [math]\displaystyle{ p_x(x,y) = \textstyle \sum\limits_{i=1}^3 \sum\limits_{j=0}^3 \frac{a_{ij} i x^{i-1} y^j}{\Delta x}, }[/math] [math]\displaystyle{ p_y(x,y) = \textstyle \sum\limits_{i=0}^3 \sum\limits_{j=1}^3 \frac{a_{ij} x^i j y^{j-1}}{\Delta y}, }[/math] [math]\displaystyle{ p_{xy}(x,y) = \textstyle \sum\limits_{i=1}^3 \sum\limits_{j=1}^3 \frac{a_{ij} i x^{i-1} j y^{j-1}}{\Delta x \Delta y}, }[/math] where [math]\displaystyle{ \Delta x }[/math] is the [math]\displaystyle{ x }[/math] spacing of the cell containing the point [math]\displaystyle{ (x,y) }[/math] and similar for [math]\displaystyle{ \Delta y }[/math]. In this case, the most practical approach to computing the coefficients [math]\displaystyle{ \alpha }[/math] is to let [math]\displaystyle{ x=\left[\begin{smallmatrix}f(0,0)&f(1,0)&f(0,1)&f(1,1)&\Delta x f_x(0,0)&\Delta xf_x(1,0)&\Delta x f_x(0,1)&\Delta x f_x(1,1)&\Delta y f_y(0,0)&\Delta y f_y(1,0)&\Delta y f_y(0,1)&\Delta y f_y(1,1)&\Delta x \Delta y f_{xy}(0,0)&\Delta x \Delta y f_{xy}(1,0)&\Delta x \Delta y f_{xy}(0,1)&\Delta x \Delta y f_{xy}(1,1)\end{smallmatrix}\right]^T, }[/math] then to solve [math]\displaystyle{ \alpha=A^{-1}x }[/math] with [math]\displaystyle{ A }[/math] as before. Next, the normalized interpolating variables are computed as [math]\displaystyle{ \begin{align} \overline{x} &= \frac{x-x_0}{x_1-x_0}, \\ \overline{y} &= \frac{y-y_0}{y_1-y_0} \end{align} }[/math] where [math]\displaystyle{ x_0, x_1, y_0, }[/math] and [math]\displaystyle{ y_1 }[/math] are the [math]\displaystyle{ x }[/math] and [math]\displaystyle{ y }[/math] coordinates of the grid points surrounding the point [math]\displaystyle{ (x,y) }[/math]. Then, the interpolating surface becomes [math]\displaystyle{ p(x,y) = \sum\limits_{i=0}^3 \sum_{j=0}^3 a_{ij} {\overline{x}}^i {\overline{y}}^j. }[/math]
Finding derivatives from function values
If the derivatives are unknown, they are typically approximated from the function values at points neighbouring the corners of the unit square, e.g. using finite differences.
To find either of the single derivatives, [math]\displaystyle{ f_x }[/math] or [math]\displaystyle{ f_y }[/math], using that method, find the slope between the two surrounding points in the appropriate axis. For example, to calculate [math]\displaystyle{ f_x }[/math] for one of the points, find [math]\displaystyle{ f(x,y) }[/math] for the points to the left and right of the target point and calculate their slope, and similarly for [math]\displaystyle{ f_y }[/math].
To find the cross derivative [math]\displaystyle{ f_{xy} }[/math], take the derivative in both axes, one at a time. For example, one can first use the [math]\displaystyle{ f_x }[/math] procedure to find the [math]\displaystyle{ x }[/math] derivatives of the points above and below the target point, then use the [math]\displaystyle{ f_y }[/math] procedure on those values (rather than, as usual, the values of [math]\displaystyle{ f }[/math] for those points) to obtain the value of [math]\displaystyle{ f_{xy}(x,y) }[/math] for the target point. (Or one can do it in the opposite direction, first calculating [math]\displaystyle{ f_y }[/math] and then [math]\displaystyle{ f_x }[/math] from those. The two give equivalent results.)
At the edges of the dataset, when one is missing some of the surrounding points, the missing points can be approximated by a number of methods. A simple and common method is to assume that the slope from the existing point to the target point continues without further change, and using this to calculate a hypothetical value for the missing point.
Bicubic convolution algorithm
Bicubic spline interpolation requires the solution of the linear system described above for each grid cell. An interpolator with similar properties can be obtained by applying a convolution with the following kernel in both dimensions: [math]\displaystyle{ W(x) = \begin{cases} (a+2)|x|^3-(a+3)|x|^2+1 & \text{for } |x| \leq 1, \\ a|x|^3-5a|x|^2+8a|x|-4a & \text{for } 1 \lt |x| \lt 2, \\ 0 & \text{otherwise}, \end{cases} }[/math] where [math]\displaystyle{ a }[/math] is usually set to −0.5 or −0.75. Note that [math]\displaystyle{ W(0)=1 }[/math] and [math]\displaystyle{ W(n)=0 }[/math] for all nonzero integers [math]\displaystyle{ n }[/math].
This approach was proposed by Keys, who showed that [math]\displaystyle{ a=-0.5 }[/math] produces third-order convergence with respect to the sampling interval of the original function.[1]
If we use the matrix notation for the common case [math]\displaystyle{ a = -0.5 }[/math], we can express the equation in a more friendly manner: [math]\displaystyle{ p(t) = \tfrac{1}{2} \begin{bmatrix} 1 & t & t^2 & t^3 \end{bmatrix} \begin{bmatrix} 0 & 2 & 0 & 0 \\ -1 & 0 & 1 & 0 \\ 2 & -5 & 4 & -1 \\ -1 & 3 & -3 & 1 \end{bmatrix} \begin{bmatrix} f_{-1} \\ f_0 \\ f_1 \\ f_2 \end{bmatrix} }[/math] for [math]\displaystyle{ t }[/math] between 0 and 1 for one dimension. Note that for 1-dimensional cubic convolution interpolation 4 sample points are required. For each inquiry two samples are located on its left and two samples on the right. These points are indexed from −1 to 2 in this text. The distance from the point indexed with 0 to the inquiry point is denoted by [math]\displaystyle{ t }[/math] here.
For two dimensions first applied once in [math]\displaystyle{ x }[/math] and again in [math]\displaystyle{ y }[/math]: [math]\displaystyle{ \begin{align} b_{-1}&= p(t_x, f_{(-1,-1)}, f_{(0,-1)}, f_{(1,-1)}, f_{(2,-1)}), \\[1ex] b_{0} &= p(t_x, f_{(-1,0)}, f_{(0,0)}, f_{(1,0)}, f_{(2,0)}), \\[1ex] b_{1} &= p(t_x, f_{(-1,1)}, f_{(0,1)}, f_{(1,1)}, f_{(2,1)}), \\[1ex] b_{2} &= p(t_x, f_{(-1,2)}, f_{(0,2)}, f_{(1,2)}, f_{(2,2)}), \end{align} }[/math] [math]\displaystyle{ p(x,y) = p(t_y, b_{-1}, b_{0}, b_{1}, b_{2}). }[/math]
Use in computer graphics
The bicubic algorithm is frequently used for scaling images and video for display (see bitmap resampling). It preserves fine detail better than the common bilinear algorithm.
However, due to the negative lobes on the kernel, it causes overshoot (haloing). This can cause clipping, and is an artifact (see also ringing artifacts), but it increases acutance (apparent sharpness), and can be desirable.
See also
- Spatial anti-aliasing
- Bézier surface
- Bilinear interpolation
- Cubic Hermite spline, the one-dimensional analogue of bicubic spline
- Lanczos resampling
- Natural neighbor interpolation
- Sinc filter
- Spline interpolation
- Tricubic interpolation
- Directional Cubic Convolution Interpolation
References
- ↑ R. Keys (1981). "Cubic convolution interpolation for digital image processing". IEEE Transactions on Acoustics, Speech, and Signal Processing 29 (6): 1153–1160. doi:10.1109/TASSP.1981.1163711. Bibcode: 1981ITASS..29.1153K.
External links
- Application of interpolation to elevation samples
- Interpolation theory
- Explanation and Java/C++ implementation of (bi)cubic interpolation
- Excel Worksheet Function for Bicubic Lagrange Interpolation
 |