Biology:H3K36me2
H3K36me2 is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the di-methylation at the 36th lysine residue of the histone H3 protein.
There are diverse modifications at H3K36 and have many important biological processes. H3K36 has different acetylation and methylation states with no similarity to each other.[1]
Nomenclature
H3K36me2 indicates dimethylation of lysine 36 on histone H3 protein subunit: [2]
| Abbr. | Meaning |
| H3 | H3 family of histones |
| K | standard abbreviation for lysine |
| 36 | position of amino acid residue
(counting from N-terminus) |
| me | methyl group |
| 2 | number of methyl groups added |
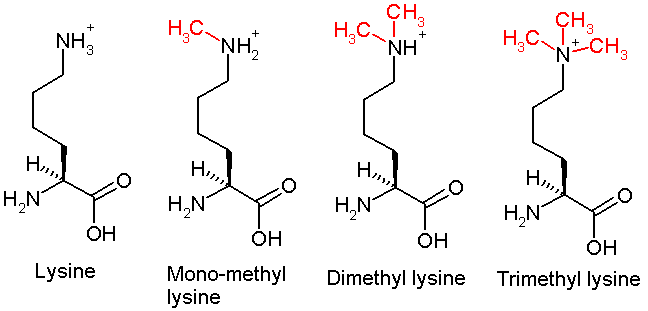
Lysine Methylation
This diagram shows the progressive methylation of a lysine residue. The di-methylation denotes the methylation present in H3K36me2.
Understanding histone modifications
The genomic DNA of eukaryotic cells is wrapped around special protein molecules known as Histones. The complexes formed by the looping of the DNA are known as chromatin. The basic structural unit of chromatin is the nucleosome: this consists of the core octamer of histones (H2A, H2B, H3 and H4) as well as a linker histone and about 180 base pairs of DNA. These core histones are rich in lysine and arginine residues. The carboxyl (C) terminal end of these histones contribute to histone-histone interactions, as well as histone-DNA interactions. The amino (N) terminal charged tails are the site of the post-translational modifications, such as the one seen in H3K36me3.[3][4]
Epigenetic implications
The post-translational modification of histone tails by either histone modifying complexes or chromatin remodelling complexes are interpreted by the cell and lead to complex, combinatorial transcriptional output. It is thought that a Histone code dictates the expression of genes by a complex interaction between the histones in a particular region.[5] The current understanding and interpretation of histones comes from two large scale projects: ENCODE and the Epigenomic roadmap.[6] The purpose of the epigenomic study was to investigate epigenetic changes across the entire genome. This led to chromatin states which define genomic regions by grouping the interactions of different proteins and/or histone modifications together. Chromatin states were investigated in Drosophila cells by looking at the binding location of proteins in the genome. Use of ChIP-sequencing revealed regions in the genome characterised by different banding.[7] Different developmental stages were profiled in Drosophila as well, an emphasis was placed on histone modification relevance.[8] A look in to the data obtained led to the definition of chromatin states based on histone modifications.[9] Certain modifications were mapped and enrichment was seen to localize in certain genomic regions. Five core histone modifications were found with each respective one being linked to various cell functions.
- H3K4me3-promoters
- H3K4me1- primed enhancers
- H3K36me3-gene bodies
- H3K27me3-polycomb repression
- H3K9me3-heterochromatin
The human genome was annotated with chromatin states. These annotated states can be used as new ways to annotate a genome independently of the underlying genome sequence. This independence from the DNA sequence enforces the epigenetic nature of histone modifications. Chromatin states are also useful in identifying regulatory elements that have no defined sequence, such as enhancers. This additional level of annotation allows for a deeper understanding of cell specific gene regulation.[10]
Methods
The histone mark H3K36me2 can be detected in a variety of ways:
1. Chromatin Immunoprecipitation Sequencing (ChIP-sequencing) measures the amount of DNA enrichment once bound to a targeted protein and immunoprecipitated. It results in good optimization and is used in vivo to reveal DNA-protein binding occurring in cells. ChIP-Seq can be used to identify and quantify various DNA fragments for different histone modifications along a genomic region.[11]
2. Micrococcal Nuclease sequencing (MNase-seq) is used to investigate regions that are bound by well positioned nucleosomes. Use of the micrococcal nuclease enzyme is employed to identify nucleosome positioning. Well positioned nucleosomes are seen to have enrichment of sequences.[12]
3. Assay for transposase accessible chromatin sequencing (ATAC-seq) is used to look in to regions that are nucleosome free (open chromatin). It uses hyperactive Tn5 transposon to highlight nucleosome localisation.[13][14][15]
See also
References
- ↑ "H3K36". https://epigenie.com/key-epigenetic-players/histone-proteins-and-modifications/histone-h3k36/. Retrieved 10 November 2019.
- ↑ Huang, Suming; Litt, Michael D.; Ann Blakey, C. (2015-11-30). Epigenetic Gene Expression and Regulation. Elsevier Science. pp. 21–38. ISBN 9780127999586.
- ↑ "Multivalent engagement of chromatin modifications by linked binding modules". Nature Reviews. Molecular Cell Biology 8 (12): 983–94. December 2007. doi:10.1038/nrm2298. PMID 18037899.
- ↑ "Chromatin modifications and their function". Cell 128 (4): 693–705. February 2007. doi:10.1016/j.cell.2007.02.005. PMID 17320507.
- ↑ "Translating the histone code". Science 293 (5532): 1074–80. August 2001. doi:10.1126/science.1063127. PMID 11498575.
- ↑ "Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project". Nature 447 (7146): 799–816. June 2007. doi:10.1038/nature05874. PMID 17571346. Bibcode: 2007Natur.447..799B.
- ↑ "Systematic protein location mapping reveals five principal chromatin types in Drosophila cells". Cell 143 (2): 212–24. October 2010. doi:10.1016/j.cell.2010.09.009. PMID 20888037.
- ↑ "Identification of functional elements and regulatory circuits by Drosophila modENCODE". Science 330 (6012): 1787–97. December 2010. doi:10.1126/science.1198374. PMID 21177974. Bibcode: 2010Sci...330.1787R.
- ↑ "Comprehensive analysis of the chromatin landscape in Drosophila melanogaster". Nature 471 (7339): 480–5. March 2011. doi:10.1038/nature09725. PMID 21179089. Bibcode: 2011Natur.471..480K.
- ↑ "Integrative analysis of 111 reference human epigenomes". Nature 518 (7539): 317–30. February 2015. doi:10.1038/nature14248. PMID 25693563. Bibcode: 2015Natur.518..317..
- ↑ "Whole-Genome Chromatin IP Sequencing (ChIP-Seq)". https://www.illumina.com/Documents/products/datasheets/datasheet_chip_sequence.pdf. Retrieved 23 October 2019.
- ↑ "MAINE-Seq/Mnase-Seq". https://www.illumina.com/science/sequencing-method-explorer/kits-and-arrays/maine-seq-mnase-seq-nucleo-seq.html?langsel=/us/. Retrieved 23 October 2019.
- ↑ Buenrostro, Jason D.; Wu, Beijing; Chang, Howard Y.; Greenleaf, William J. (2015). "ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide". Current Protocols in Molecular Biology 109: 21.29.1–21.29.9. doi:10.1002/0471142727.mb2129s109. ISBN 9780471142720. PMID 25559105.
- ↑ Schep, Alicia N.; Buenrostro, Jason D.; Denny, Sarah K.; Schwartz, Katja; Sherlock, Gavin; Greenleaf, William J. (2015). "Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions". Genome Research 25 (11): 1757–1770. doi:10.1101/gr.192294.115. ISSN 1088-9051. PMID 26314830.
- ↑ Song, L.; Crawford, G. E. (2010). "DNase-seq: A High-Resolution Technique for Mapping Active Gene Regulatory Elements across the Genome from Mammalian Cells". Cold Spring Harbor Protocols 2010 (2): pdb.prot5384. doi:10.1101/pdb.prot5384. ISSN 1559-6095. PMID 20150147.
 |