Biology:F-ATPase
| F-ATPase | |
|---|---|
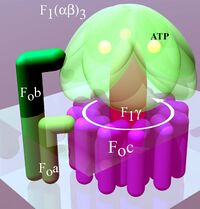 Simplified model of FOF1-ATPase alias ATP synthase of E. coli. Subunits of the enzyme are labeled accordingly | |
| Identifiers | |
| Symbol | F-ATPase |
| TCDB | 3.A.2 |
| OPM superfamily | 5 |
| OPM protein | 6fkf |
| Membranome | 227 |
F-ATPase, also known as F-Type ATPase, is an ATPase/synthase found in bacterial plasma membranes, in mitochondrial inner membranes (in oxidative phosphorylation, where it is known as Complex V), and in chloroplast thylakoid membranes. It uses a proton gradient to drive ATP synthesis by allowing the passive flux of protons across the membrane down their electrochemical gradient and using the energy released by the transport reaction to release newly formed ATP from the active site of F-ATPase. Together with V-ATPases and A-ATPases, F-ATPases belong to superfamily of related rotary ATPases.
F-ATPase consists of two domains:
- the Fo domain, which is integral in the membrane and is composed of 3 different types of integral proteins classified as a, b and c.[1]
- the F1, which is peripheral (on the side of the membrane that the protons are moving into). F1 is composed of 5 polypeptide units α3β3γδε that bind to the surface of the Fo domain.[1]
F-ATPases usually work as ATP synthases instead of ATPases in cellular environments. That is to say, it usually makes ATP from the proton gradient instead of working in the other direction like V-ATPases typically do. They do occasionally revert as ATPases in bacteria.[2]
Structure
Fo-F1 particles are mainly formed of polypeptides. The F1-particle contains 5 types of polypeptides, with the composition-ratio—3α:3β:1δ:1γ:1ε. The Fo has the 1a:2b:12c composition. Together they form a rotary motor. As the protons bind to the subunits of the Fo domains, they cause parts of it to rotate. This rotation is propagated by a 'camshaft' to the F1 domain. ADP and Pi (inorganic phosphate) bind spontaneously to the three β subunits of the F1 domain, so that every time it goes through a 120° rotation ATP is released (rotational catalysis).
The Fo domains sits within the membrane, spanning the phospholipid bilayer, while the F1 domain extends into the cytosol of the cell to facilitate the use of newly synthesized ATP.
The Bovine Mitochondrial F1-ATPase Complexed with the inhibitor protein If1 is commonly cited in the relevant literature. Examples of its use may be found in many cellular fundamental metabolic activities such as acidosis and alkalosis and respiratory gas exchange.
The o in the Fo stands for oligomycin, because oligomycin is able to inhibit its function.
N-ATPase
N-ATPases are a group of F-type ATPases without a delta/OSCP subunit, found in bacteria and a group of archaea via horizontal gene transfer. They transport sodium ions instead of protons and tend to hydrolyze ATP. They form a distinct group that is further apart from usual F-ATPases than A-ATPases are from V-ATPases.[3]
References
- ↑ 1.0 1.1 Lodish, Harvey (2008). Molecular Cell Biology (Kindle ed.). W. H. Freeman. p. 553. https://archive.org/details/molecularcellbio00harv_624.
- ↑ "Transport, Solute" (in en). Encyclopedia of Microbiology (Third ed.). Academic Press. 2009. pp. 529–544. doi:10.1016/B978-012373944-5.00107-3. ISBN 978-0-12-373944-5. https://archive.org/details/deskencyclopedia00scha.
- ↑ Dibrova, DV; Galperin, MY; Mulkidjanian, AY (15 June 2010). "Characterization of the N-ATPase, a distinct, laterally transferred Na+-translocating form of the bacterial F-type membrane ATPase.". Bioinformatics 26 (12): 1473–6. doi:10.1093/bioinformatics/btq234. PMID 20472544.
 |

