Biology:Plasmodium falciparum erythrocyte membrane protein 1
| PfEMP1, N-terminal segment | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | NTS | ||||||||
| Pfam | PF15447 | ||||||||
| InterPro | IPR029210 | ||||||||
| |||||||||
| PfEMP DBL domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | PFEMP | ||||||||
| Pfam | PF03011 | ||||||||
| InterPro | IPR004258 | ||||||||
| CATH | 2yk0A03 | ||||||||
| |||||||||
| PfEMP CIDR1γ | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | CIDR1_gamma | ||||||||
| Pfam | PF18562 | ||||||||
| InterPro | IPR041480 | ||||||||
| |||||||||
| PfEMP1, acidic terminal segment | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | ATS | ||||||||
| Pfam | PF15445 | ||||||||
| InterPro | IPR029211 | ||||||||
| |||||||||
Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) is a family of proteins present on the membrane surface of red blood cells (RBCs or erythrocytes) that are infected by the malarial parasite Plasmodium falciparum. PfEMP1 is synthesized during the parasite's blood stage (erythrocytic schizogony) inside the RBC, during which the clinical symptoms of falciparum malaria are manifested. Acting as both an antigen and adhesion protein, it is thought to play a key role in the high level of virulence associated with P. falciparum. It was discovered in 1984 when it was reported that infected RBCs had unusually large-sized cell membrane proteins, and these proteins had antibody-binding (antigenic) properties. An elusive protein, its chemical structure and molecular properties were revealed only after a decade, in 1995. It is now established that there is not one but a large family of PfEMP1 proteins, genetically regulated (encoded) by a group of about 60 genes called var. Each P. falciparum is able to switch on and off specific var genes to produce a functionally different protein, thereby evading the host's immune system. RBCs carrying PfEMP1 on their surface stick to endothelial cells, which facilitates further binding with uninfected RBCs (through the processes of sequestration and rosetting), ultimately helping the parasite to both spread to other RBCs as well as bringing about the fatal symptoms of P. falciparum malaria.
Introduction
Malaria is the deadliest among infectious diseases, accounting for approximately 429,000 human deaths in 2015 as of the latest estimate by the World Health Organization.[2] In humans, malaria can be caused by five Plasmodium parasites, namely P. falciparum, P. vivax, P. malariae, P. ovale and P. knowlesi.[3] P. falciparum is the most dangerous species, attributed to >99% of malaria's death toll, with 70% of these deaths occurring in children under the age of five years.[2] The parasites are transmitted through the bites of female mosquitos (of the species of Anopheles). Before invading the RBCs and causing the symptoms of malaria, the parasites first multiply in the liver. The daughter parasites called merozoites then only infect the RBCs. They undergo structural development inside the RBCs, becoming trophozoites and schizonts. It is during this period that malarial symptoms are produced.[4]
Unlike RBCs infected by other Plasmodium species, P. falciparum-infected RBCs had been known to spontaneously stick together. By the early 1980s, it was established that when the parasite (both the trophozoite and schizont forms) enters the blood stream and infects RBCs, the infected cells form knobs on their surface. Then they become sticky, and get attached to the walls (endothelium) of the blood vessels through a process called cytoadhesion, or cytoadherence.[5] Such attachment favours binding with and accumulation of other RBCs. This process is known as sequestration.[6] It is during this condition that the parasites induce an immune response (antigen-antibody reaction) and evade destruction in the spleen.[7][8] Although the process and significance of sequestration were described in detail by two Italian physicians Amico Bignami and Ettore Marchiafava in the early 1890s, it took a century to discover the actual factor for the stickiness and virulence.[9][10]
Discovery
PfEMP1 was discovered by Russell J. Howard and his colleagues at the US National Institutes of Health in 1984. Using the techniques of radioiodination and immunoprecipitation, they found a unique but yet unknown antigen from P. falciparum-infected RBCs that appeared to cause binding with other cells.[11] Since the antigenic protein could only be detected in infected cells, they asserted that the protein was produced by the malarial parasite, and not by RBCs. The antigen was large and appeared to be different in size in different strains of P. falciparum obtained from night monkey (Aotus). In one strain, called Camp (from Malaysia), the antigen was found to have a molecular size of approximately 285 kDa; while in the other, called St. Lucia (from El Salvador), it was approximately 260 kDa. Both antigens bind to cultured skin cancer (melanoma) cells. But the researchers failed to confirm whether or not the protein actually was an adhesion molecule to the wall of blood vessels.[12] Later in the same year, they found out that the unknown antigen was associated only with RBCs having small lumps called knobs on their surface.[13] The first human RBC antigen was reported in 1986. Howard's team found that the antigens from Gambian children, who were suffering from falciparum malaria, were similar to those from the RBCs of night monkey. They determined that the molecular sizes of the proteins ranged from 250 to 300 kDa.[14]
In 1987, they discovered another type of surface antigen from the same Camp and St. Lucia strains of malarial parasites. This was also a large-sized protein of about 300 kDa, but quite different from the antigens reported in 1984. The new protein was unable to bind to melanoma cells and present only inside the cell. Hence, they named the earlier protein Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1), to distinguish it from the newly identified Plasmodium falciparum erythrocyte membrane protein 2 (PfEMP2).[15] The distinction was confirmed the next year, with an additional information that PfEMP1 is relatively less in number.[16]
Although some of the properties of PfEMP1 were firmly established, the protein was difficult to isolate due to its low occurrence. Five years after its discovery, one of the original researchers Irwin Sherman began to doubt the existence of PfEMP1 as a unique protein.[10] He argued that the antigen could be merely a surface protein of RBCs that changes upon infection with malarial parasites.[17] A consensus was achieved in 1995 following the identification (by cloning) of the gene for PfEMP1. The discovery of the genes was independently reported by Howard's team and two other teams at NIH. Howard's team identified two genes for PfEMP1, and recombinant protein products of these genes were shown to have antigenic and adhesive properties. They further affirmed that PfEMP1 is the key molecule in the ability of P. falciparum to evade the host's immune system.[18] Joseph D. Smith and others showed that PfEMP1 is actually a large family of proteins encoded by a multigene family called var. The gene products can bind to a variety of receptors including those on endothelial cells.[19] Xin-Zhuan Su and others showed that there could be more than 50 var genes which are distributed on different chromosomes of the malarial parasite.[20]
Structure
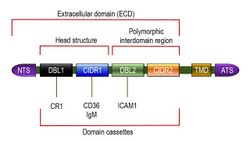
PfEMP1 is a large family of proteins having high molecular weights ranging from 200 to 350 kDa.[21] The wide range of molecular size reflects extreme variation in the amino acid composition of the proteins. But all the PfEMP1 proteins can be described as having three basic structural components, namely, an extracellular domain (ECD), a transmembrane domain (TMD) and an intracellular acidic terminal segment (ATS). The extracellular domain is fully exposed on the cell surface, and is the most variable region. It consists of a number of sub-domains, including a short and conserved N terminal segment (NTS) at the outermost region, followed by a highly variable Duffy-binding-like (DBL) domain, sometimes a Ca2+-binding C2 domain, and then one or two cysteine-rich interdomain regions (CIDRs).[9][22]
Duffy-binding-like domains are so named because of their similarity to the Duffy binding proteins of P. vivax and P. knowlesi.[23] There are six variant types of DBL, named DBLα, DBLβ, DBLγ, DBLδ, DBLε and DBLζ. CIDR is also divided into three classes: CIDRα, CIDRβ and CIDRγ.[24] Both DBL and CIDR have an additional type called PAM, so named because of their specific involvement in pregnancy-associated malaria (PAM).[25] In spite of the diverse DBL and CIDR proteins, the extracellular amino terminal region is partly conserved, consisting of about 60 amino acids of NTS, one each of DBLα and CIDR1 proteins in tandem. This semi-conserved DBLα-CIDR1 region is called the head structure.[26] The last CIDR region joins the TMD, which is embedded in the cell membrane. The TMD and ATS are highly conserved among different PfEMP1s,[27] and their structures have been solved using solution NMR (PDB: 2LKL).[28]
The head structure is followed by a variable combination of diverse DBL and CIDR proteins, and in many cases along with C2. This variation gives rise to different types of PfEMP1. The DBL-CIDR combination in a particular type of PfEMP1 protein is never random, but organized into specific sequences known as domain cassettes.[29] In some domain cassettes, there are only two or few DBL domains and CIDR domains, but in others they cover the entire length of the PfEMP1. These differences are responsible for different binding capacity among different PfEMP1s.[30] For instance, among the most well-known types, VAR3 (earlier called type 3 PfEMP1) is the smallest, consisting of only NTS with DBL1α and DBL2ε domains in the ECD. Its molecular size is approximately 150 kDa.[31] In domain cassette (DC) 4 type, the ECD is made up of three domains DBLα1.1/1.4, CIDRα1.6 and DBLβ3. The DBLβ3 domain contains a binding site for intercellular adhesion molecule 1 (ICAM1). This is particularly implicated with the development of brain infection.[32] VAR2CSA is atypical in having a single domain cassette that consists of three N terminal DBLPAM domains followed by three DBLε domains and one CIDRPAM. The seven domains always occur together. The usual NTS is absent.[33][34] The protein specifically binds to chondroitin sulphate A (CSA); hence the name VAR2CSA.[35]
Synthesis and transport
The PfEMP1 proteins are regulated and produced (encoded) by about 60 different var genes,[37] but an individual P. falciparum would switch on only a single var gene at a time to produce only one type of PfEMP.[38][39] The var genes are distributed in two exons. Exon 1 encodes amino acids of the highly variable ECD,[40] while exon 2 encodes those of the conserved TMD and ATS.[41] Based on their location in the chromosome and sequence, the var genes are generally classified into three major groups, A, B, and C, and two intermediate groups, B/A and B/C;[9][42] or sometimes simply into five classes, upsA, upsB, upsC, upsD, and upsE respectively.[43] Groups A and B are found towards the terminal end (subtelomeric) region of the chromosome, while group C is in the central (centromeric) region.[44][45]
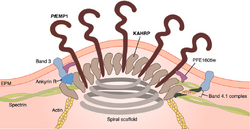
Once the PfEMP1 protein is fully synthesized (translated), it is carried to the cytoplasm towards the RBC membrane. The NTS is crucial for such directional movement.[26] Within the cytoplasm, the newly synthesized protein is attached to a Golgi-like membranous vesicle called the Maurer's cleft.[46] Inside the Maurer's clefts is a family of proteins called Plasmodium helical interspersed subtelomeric (PHIST) proteins. Of the PHIST proteins, PFI1780w and PFE1605w bind the intracellular ATS of PfEMP1 during transport to the RBC membrane.[28][47]
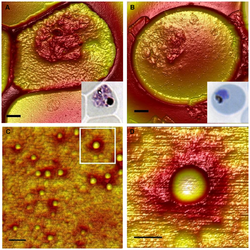
The PfEMP1 molecule is deposited at the RBC membrane at the knobs.[48] These knobs are easily identified as conspicuous bumps on the infected RBCs from the early trophozoite stage onward.[49] The malarial parasite cannot induce its virulence on RBCs without knobs.[50] As many as 10,000 knobs are distributed throughout the surface of a mature infected RBC, and each knob is 50-80 nm in diameter.[1] The export of pfEMP1 from Maurer's cleft to RBC membrane is mediated by binding of another protein produced by the parasite called knob-associated histidine-rich protein (KAHRP). KAHRP enhances the structural rigidity of infected RBC and adhesion of PfEMP1 on the knobs.[51] It is also directly responsible for forming knobs, as indicated by the fact that kahrp gene-deficient malarial parasites do not form knobs.[52] To form a knob, KAHRP aggregates several membrane skeletal proteins of the host RBC, such as spectrin, actin, ankyrin R, and spectrin–actin band 4.1 complex.[53] Upon arrival at the knob, PfEMP1 is attached to the spectrin network using the PHIST proteins.[54][36]
Function
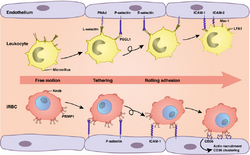
The primary function of PfEMP1 is to bind and attach RBCs to the wall of the blood vessels. The most important binding properties of P. falciparum known to date are mediated by the head structure of PfEMP1, consisting of DBL domains and CIDRs.[55] DBL domains can bind to a variety of cell receptors including thrombospondin (TSP), complement receptor 1 (CR1), chondroitin sulfate A (CSA),[5] P-selectin,[56] endothelial protein C receptor (EPCR),[57] and heparan sulfate.[58] The DBL domain adjacent to the head structure binds to ICAM-1.[59] CIDRs mainly bind to a large variety of cluster determinant 36 (CD36).[21][60] These bindings produce the pathogenic characteristics of the parasite, such as sequestration of infected cells in different tissues,[61] invasion of RBCs,[62] and clustering of infected cells by a process called rosetting.[63]

CIDR1 protein in the semi-conserved head structure is the principal and best understood adhesion site of PfEMP1. It binds with CD36 on endothelial cells.[65][66] Only group B and C proteins are able to bind, and that too with only those having CIDRα2-6 sequence types. On the other hand, group A proteins have either CIDRα1 or CIDRβ/γ/δ, and they are responsible for the most severe condition of malaria.[45] Binding with ICAM-1 is achieved through the DBLβ domain adjacent to the head structure. However, many PfEMP1s having DBLβ domain do not bind to ICAM-1,[67] and it appears that only the DBLβ paired with C2 domain can to bind to ICAM-1.[61] The DBLα-CIDRγ tandem pair is the main factor for rosetting,[64] sticking together the infected RBC with the uninfected cells, and thereby clogging of the blood vessels. This activity is performed through binding with CR1.[63][68]
The most dangerous malarial infection is in the brain and is called cerebral malaria. In cerebral malaria, the PfEMP1 proteins involved are DC8 and DC13. They are named after the number of domain cassettes they contain, and are capable of binding not only endothelial cells of the brain, but also in different organs including brain, lung, heart, and bone marrow.[69] Initially, it was assumed that PfEMP1 binds to ICAM-1 in the brain, but DC8 and DC13 were found incompatible with ICAM-1. Instead DC8 and DC13 specifically bind to EPCR using CIDRα sub-types such as CIDRα1.1, CIDRα1.4, CIDRα1.5 and CIDRα1.7.[57] However, it was later shown that DC13 can bind to both ICAM-1 and EPCR.[70] EPCR is thus a potential vaccine and drug target in cerebral malaria.[71]
VAR2CSA is unique in that it is mostly produced by the placenta during pregnancy (the condition called pregnancy-associated malaria, PAM, or placental malaria). The majority of PAM is therefore due to VAR2SCA.[27] Unlike other PfEMP1, VAR2CSA binds to chondroitin sulfate A present on the vascular endothelium of the placenta. Although its individual domains can bind to CSA, its entire structure is used for complete binding.[72] The major complication in PAM is low-birth-weight babies. However, women who survived the first infection generally develop an effective immune response. In P. falciparum-prevalent regions in Africa, pregnant women are found to contain high levels of antibody (immunoglobulin G, or IgG) against VAR2CSA, which protect them the placenta-attacking malarial parasite. They are noted for giving birth to heavier babies.[33]
Clinical importance
In a normal human immune system, malarial parasite binding to RBCs stimulates the production of antibodies that attack the PfEMP1 molecules. Binding of antibody with PfEMP1 disables the binding properties of DBL domains, causing loss of cell adhesion, and the infected RBC is destroyed. In this scenario, malaria is avoided.[73] However, to evade the host's immune response, different P. falciparum switch on and off different var genes to produce functionally different (antigenically distinct) PfEMP1s. Each variant type of PfEMP1 has different binding property, and thus, is not always recognized by antibodies.[74]
By default, all the var genes in the malarial parasite are inactivated. Activation (gene expression) of var is initiated upon infection of the organs. Further, in each organ only specific var genes are activated. The severity of the infection is determined by the type of organ in which infection occurs, hence, the type of var gene activated. For examples, in the most severe cases of malaria, such as cerebral malaria, only the var genes for the PfEMP1 proteins DC8 and DC13 are switched on.[75][76] Upon the synthesis of DC8 and DC13, their CIDRα1 domains bind to EPCR, which brings about the onset of severe malaria.[77] The abundance of the gene products (transcripts) of these PfEMP1 proteins (specifically the CIDRα1 subtype transcripts) directly relates to the severity of the disease. This further indicates that preventing the interaction between CIDRα1 and EPCR would be good target for a potential vaccine.[78][79] In pregnancy-associated malaria, another severe type of falciparum malaria, the gene for VAR2CSA (named var2csa) is activated in the placenta. Binding of VAR2CSA to CSA is the primary cause of premature delivery, death of the foetus and severe anaemia in the mother.[72] This indicates that drugs targeting VAR2CSA will be able to prevent the effects of malaria, and for this reason VAR2CSA is the leading candidate for development of a PAM vaccine.[80]
References
- ↑ 1.0 1.1 "The density of knobs on Plasmodium falciparum-infected erythrocytes depends on developmental age and varies among isolates". PLOS ONE 7 (9): e45658. 2012. doi:10.1371/journal.pone.0045658. PMID 23029166. Bibcode: 2012PLoSO...745658Q.
- ↑ 2.0 2.1 "Malaria Fact sheet". WHO Media Centre. April 2016. https://www.who.int/mediacentre/factsheets/fs094/en/.
- ↑ "History of the discovery of the malaria parasites and their vectors". Parasites & Vectors 3 (1): 5. February 2010. doi:10.1186/1756-3305-3-5. PMID 20205846.
- ↑ "Biology of human malaria plasmodia including Plasmodium knowlesi". Mediterranean Journal of Hematology and Infectious Diseases 4 (1): e2012013. 2012. doi:10.4084/MJHID.2012.013. PMID 22550559.
- ↑ 5.0 5.1 "Adhesion of Plasmodium falciparum-infected erythrocytes to human cells: molecular mechanisms and therapeutic implications". Expert Reviews in Molecular Medicine 11: e16. May 2009. doi:10.1017/S1462399409001082. PMID 19467172.
- ↑ "Knobs, knob proteins and cytoadherence in falciparum malaria". The International Journal of Biochemistry 23 (9): 775–89. 1991. doi:10.1016/0020-711X(91)90061-Q. PMID 1773882.
- ↑ "Parasite sequestration in Plasmodium falciparum malaria: spleen and antibody modulation of cytoadherence of infected erythrocytes". Proceedings of the National Academy of Sciences of the United States of America 80 (16): 5075–9. August 1983. doi:10.1073/pnas.80.16.5075. PMID 6348780. Bibcode: 1983PNAS...80.5075D.
- ↑ "Sequestration in Plasmodium falciparum malaria: sticky cells and sticky problems". Parasitology Today 6 (8): 247–54. August 1990. doi:10.1016/0169-4758(90)90184-6. PMID 15463355.
- ↑ 9.0 9.1 9.2 "The role of PfEMP1 as targets of naturally acquired immunity to childhood malaria: prospects for a vaccine". Parasitology 143 (2): 171–86. February 2016. doi:10.1017/S0031182015001274. PMID 26741401.
- ↑ 10.0 10.1 Sherman, Irwin (2008). Advances in Parasitology: Reflections on a Century of Malaria Biochemistry. London (UK): Academic Press. pp. 188–189. ISBN 978-0-08-092183-9. https://books.google.com/books?id=t9e81NrD0FIC.
- ↑ "Malaria, the red cell, and the endothelium". Annual Review of Medicine 45 (1): 283–95. 1994. doi:10.1146/annurev.med.45.1.283. PMID 8198384.
- ↑ "Identification of a strain-specific malarial antigen exposed on the surface of Plasmodium falciparum-infected erythrocytes". The Journal of Experimental Medicine 159 (6): 1567–75. June 1984. doi:10.1084/jem.159.6.1567. PMID 6374009.
- ↑ "Knob-positive and knob-negative Plasmodium falciparum differ in expression of a strain-specific malarial antigen on the surface of infected erythrocytes". The Journal of Experimental Medicine 160 (5): 1585–90. November 1984. doi:10.1084/jem.160.5.1585. PMID 6208311.
- ↑ "Identification of isolate-specific proteins on sorbitol-enriched Plasmodium falciparum infected erythrocytes from Gambian patients". Parasitology 92 ( Pt 3) (3): 511–25. June 1986. doi:10.1017/S0031182000065410. PMID 3526259.
- ↑ "Transport of an Mr approximately 300,000 Plasmodium falciparum protein (Pf EMP 2) from the intraerythrocytic asexual parasite to the cytoplasmic face of the host cell membrane". The Journal of Cell Biology 104 (5): 1269–80. May 1987. doi:10.1083/jcb.104.5.1269. PMID 2437128.
- ↑ "Two approximately 300 kilodalton Plasmodium falciparum proteins at the surface membrane of infected erythrocytes". Molecular and Biochemical Parasitology 27 (2–3): 207–23. January 1988. doi:10.1016/0166-6851(88)90040-0. PMID 3278227.
- ↑ "Antigens on the Plasmodium falciparum infected erythrocyte surface are not parasite derived". Parasitology Today 6 (10): 317–20. October 1990. doi:10.1016/0169-4758(90)90174-3. PMID 15463255.
- ↑ "Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes". Cell 82 (1): 77–87. July 1995. doi:10.1016/0092-8674(95)90054-3. PMID 7541722.
- ↑ "Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes". Cell 82 (1): 101–10. July 1995. doi:10.1016/0092-8674(95)90056-X. PMID 7606775.
- ↑ "The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes". Cell 82 (1): 89–100. July 1995. doi:10.1016/0092-8674(95)90055-1. PMID 7606788.
- ↑ 21.0 21.1 "PfEMP1: an antigen that plays a key role in the pathogenicity and immune evasion of the malaria parasite Plasmodium falciparum". The International Journal of Biochemistry & Cell Biology 41 (7): 1463–6. July 2009. doi:10.1016/j.biocel.2008.12.012. PMID 19150410.
- ↑ "var genes, PfEMP1 and the human host". Molecular and Biochemical Parasitology 134 (1): 3–9. March 2004. doi:10.1016/j.molbiopara.2003.09.010. PMID 14747137.
- ↑ "Identification of the erythrocyte binding domains of Plasmodium vivax and Plasmodium knowlesi proteins involved in erythrocyte invasion". The Journal of Experimental Medicine 180 (2): 497–506. August 1994. doi:10.1084/jem.180.2.497. PMID 8046329.
- ↑ "Classification of adhesive domains in the Plasmodium falciparum erythrocyte membrane protein 1 family". Molecular and Biochemical Parasitology 110 (2): 293–310. October 2000. doi:10.1016/S0166-6851(00)00279-6. PMID 11071284.
- ↑ "The chondroitin sulfate A-binding site of the VAR2CSA protein involves multiple N-terminal domains". The Journal of Biological Chemistry 286 (18): 15908–17. May 2011. doi:10.1074/jbc.M110.191510. PMID 21398524.
- ↑ 26.0 26.1 "Identification of a role for the PfEMP1 semi-conserved head structure in protein trafficking to the surface of Plasmodium falciparum infected red blood cells". Cellular Microbiology 12 (10): 1446–62. October 2010. doi:10.1111/j.1462-5822.2010.01481.x. PMID 20438573.
- ↑ 27.0 27.1 Hviid, Lars; Jensen, Anja T.R. (2015). "PfEMP1 – A Parasite Protein Family of Key Importance in Plasmodium falciparum Malaria Immunity and Pathogenesis". in Rollinson, D.; Stothard, J.R.. PfEMP1 – A Parasite Protein Family of Key Importance in Plasmodium falciparum Malaria Immunity and Pathogenesis. Advances in Parasitology. 88. Elsevier. pp. 51–84. doi:10.1016/bs.apar.2015.02.004. ISBN 978-0-12-802268-9.
- ↑ 28.0 28.1 "Structural analysis of the Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) intracellular domain reveals a conserved interaction epitope". The Journal of Biological Chemistry 287 (10): 7182–9. March 2012. doi:10.1074/jbc.M111.330779. PMID 22249178.
- ↑ "Global genetic diversity and evolution of var genes associated with placental and severe childhood malaria". Molecular and Biochemical Parasitology 148 (2): 169–80. August 2006. doi:10.1016/j.molbiopara.2006.03.012. PMID 16697476.
- ↑ "Plasmodium falciparum erythrocyte membrane protein 1 domain cassettes 8 and 13 are associated with severe malaria in children". Proceedings of the National Academy of Sciences of the United States of America 109 (26): E1791-800. June 2012. doi:10.1073/pnas.1120455109. PMID 22619319.
- ↑ "The var3 genes of Plasmodium falciparum 3D7 strain are differentially expressed in infected erythrocytes". Parasite 21: 19. 2014. doi:10.1051/parasite/2014019. PMID 24759654.
- ↑ "A novel domain cassette identifies Plasmodium falciparum PfEMP1 proteins binding ICAM-1 and is a target of cross-reactive, adhesion-inhibitory antibodies". Journal of Immunology 190 (1): 240–9. January 2013. doi:10.4049/jimmunol.1202578. PMID 23209327.
- ↑ 33.0 33.1 "Evidence for the involvement of VAR2CSA in pregnancy-associated malaria". The Journal of Experimental Medicine 200 (9): 1197–203. November 2004. doi:10.1084/jem.20041579. PMID 15520249.
- ↑ "VAR2CSA and protective immunity against pregnancy-associated Plasmodium falciparum malaria". Parasitology 134 (Pt 13): 1871–6. 2007. doi:10.1017/S0031182007000121. PMID 17958922. https://zenodo.org/record/999281.
- ↑ "Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria". Molecular Microbiology 49 (1): 179–91. July 2003. doi:10.1046/j.1365-2958.2003.03570.x. PMID 12823820.
- ↑ 36.0 36.1 36.2 "Modeling cytoadhesion of Plasmodium falciparum-infected erythrocytes and leukocytes-common principles and distinctive features". FEBS Letters 590 (13): 1955–71. July 2016. doi:10.1002/1873-3468.12142. PMID 26992823.
- ↑ "Genome sequence of the human malaria parasite Plasmodium falciparum". Nature 419 (6906): 498–511. October 2002. doi:10.1038/nature01097. PMID 12368864. Bibcode: 2002Natur.419..498G.
- ↑ "Developmental selection of var gene expression in Plasmodium falciparum". Nature 394 (6691): 392–5. July 1998. doi:10.1038/28660. PMID 9690477. Bibcode: 1998Natur.394..392C.
- ↑ "Antigenic variation in Plasmodium falciparum". Annual Review of Microbiology 62 (1): 445–70. 2008. doi:10.1146/annurev.micro.61.080706.093134. PMID 18785843.
- ↑ "Antigenic variation in Plasmodium falciparum: gene organization and regulation of the var multigene family". Eukaryotic Cell 6 (9): 1511–20. September 2007. doi:10.1128/EC.00173-07. PMID 17644655.
- ↑ "A well-conserved Plasmodium falciparum var gene shows an unusual stage-specific transcript pattern". Molecular Microbiology 48 (5): 1339–48. June 2003. doi:10.1046/j.1365-2958.2003.03505.x. PMID 12787360.
- ↑ "Differential var gene transcription in Plasmodium falciparum isolates from patients with cerebral malaria compared to hyperparasitaemia". Molecular and Biochemical Parasitology 150 (2): 211–8. December 2006. doi:10.1016/j.molbiopara.2006.08.005. PMID 16996149.
- ↑ "Recent advances in malaria genomics and epigenomics". Genome Medicine 8 (1): 92. September 2016. doi:10.1186/s13073-016-0343-7. PMID 27605022.
- ↑ "Plasmodium falciparum erythrocyte membrane protein 1 diversity in seven genomes--divide and conquer". PLOS Computational Biology 6 (9): e1000933. September 2010. doi:10.1371/journal.pcbi.1000933. PMID 20862303. Bibcode: 2010PLSCB...6E0933R.
- ↑ 45.0 45.1 "The role of PfEMP1 adhesion domain classification in Plasmodium falciparum pathogenesis research". Molecular and Biochemical Parasitology 195 (2): 82–7. July 2014. doi:10.1016/j.molbiopara.2014.07.006. PMID 25064606.
- ↑ "Maurer's clefts, the enigma of Plasmodium falciparum". Proceedings of the National Academy of Sciences of the United States of America 110 (50): 19987–94. December 2013. doi:10.1073/pnas.1309247110. PMID 24284172. Bibcode: 2013PNAS..11019987M.
- ↑ "Plasmodium Helical Interspersed Subtelomeric (PHIST) Proteins, at the Center of Host Cell Remodeling". Microbiology and Molecular Biology Reviews 80 (4): 905–27. December 2016. doi:10.1128/MMBR.00014-16. PMID 27582258. PMC 5116875. https://zenodo.org/record/1000776.
- ↑ "Falciparum malaria: sticking up, standing out and out-standing". Parasitology Today 16 (10): 416–20. October 2000. doi:10.1016/S0169-4758(00)01753-1. PMID 11006472.
- ↑ "Plasmodium falciparum-infected erythrocytes: qualitative and quantitative analyses of parasite-induced knobs by atomic force microscopy". Journal of Structural Biology 130 (1): 34–44. May 2000. doi:10.1006/jsbi.2000.4236. PMID 10806089. https://zenodo.org/record/1229912.
- ↑ "Exported proteins required for virulence and rigidity of Plasmodium falciparum-infected human erythrocytes". Cell 134 (1): 48–61. July 2008. doi:10.1016/j.cell.2008.04.051. PMID 18614010.
- ↑ "A spiral scaffold underlies cytoadherent knobs in Plasmodium falciparum-infected erythrocytes". Blood 127 (3): 343–51. January 2016. doi:10.1182/blood-2015-10-674002. PMID 26637786.
- ↑ "Targeted gene disruption shows that knobs enable malaria-infected red cells to cytoadhere under physiological shear stress". Cell 89 (2): 287–96. April 1997. doi:10.1016/S0092-8674(00)80207-X. PMID 9108483.
- ↑ "The role of KAHRP domains in knob formation and cytoadherence of P falciparum-infected human erythrocytes". Blood 108 (1): 370–8. July 2006. doi:10.1182/blood-2005-11-4624. PMID 16507777.
- ↑ "A Plasmodium falciparum PHIST protein binds the virulence factor PfEMP1 and comigrates to knobs on the host cell surface". FASEB Journal 28 (10): 4420–33. October 2014. doi:10.1096/fj.14-256057. PMID 24983468.
- ↑ "Plasmodium falciparum virulence determinants unveiled". Genome Biology 3 (11): REVIEWS1031. October 2002. doi:10.1186/gb-2002-3-11-reviews1031. PMID 12441004.
- ↑ "Plasmodium falciparum erythrocyte membrane protein 1 functions as a ligand for P-selectin". Blood 98 (10): 3132–5. November 2001. doi:10.1182/blood.V98.10.3132. PMID 11698301.
- ↑ 57.0 57.1 "Severe malaria is associated with parasite binding to endothelial protein C receptor". Nature 498 (7455): 502–5. June 2013. doi:10.1038/nature12216. PMID 23739325. Bibcode: 2013Natur.498..502T.
- ↑ "Binding of subdomains 1/2 of PfEMP1-DBL1α to heparan sulfate or heparin mediates Plasmodium falciparum rosetting". PLOS ONE 10 (3): e0118898. 2015. doi:10.1371/journal.pone.0118898. PMID 25742651. Bibcode: 2015PLoSO..1018898A.
- ↑ "Identification of a Plasmodium falciparum intercellular adhesion molecule-1 binding domain: a parasite adhesion trait implicated in cerebral malaria". Proceedings of the National Academy of Sciences of the United States of America 97 (4): 1766–71. February 2000. doi:10.1073/pnas.040545897. PMID 10677532. Bibcode: 2000PNAS...97.1766S.
- ↑ "A family affair: var genes, PfEMP1 binding, and malaria disease". Current Opinion in Microbiology 9 (4): 374–80. August 2006. doi:10.1016/j.mib.2006.06.006. PMID 16814594.
- ↑ 61.0 61.1 "Mapping a common interaction site used by Plasmodium falciparum Duffy binding-like domains to bind diverse host receptors". Molecular Microbiology 67 (1): 78–87. January 2008. doi:10.1111/j.1365-2958.2007.06019.x. PMID 18047571.
- ↑ "Invasion of red blood cells by malaria parasites". Cell 124 (4): 755–66. February 2006. doi:10.1016/j.cell.2006.02.006. PMID 16497586.
- ↑ 63.0 63.1 "P. falciparum rosetting mediated by a parasite-variant erythrocyte membrane protein and complement-receptor 1". Nature 388 (6639): 292–5. July 1997. doi:10.1038/40888. PMID 9230440. Bibcode: 1997Natur.388..292R.
- ↑ 64.0 64.1 "Structural basis for the ABO blood-group dependence of Plasmodium falciparum rosetting". PLOS Pathogens 8 (7): e1002781. 2012. doi:10.1371/journal.ppat.1002781. PMID 22807674.
- ↑ "Identification of a region of PfEMP1 that mediates adherence of Plasmodium falciparum infected erythrocytes to CD36: conserved function with variant sequence". Blood 90 (9): 3766–75. November 1997. doi:10.1182/blood.V90.9.3766. PMID 9345064.
- ↑ "The structural basis for CD36 binding by the malaria parasite". Nature Communications 7: 12837. September 2016. doi:10.1038/ncomms12837. PMID 27667267. Bibcode: 2016NatCo...712837H.
- ↑ "Disguising itself--insights into Plasmodium falciparum binding and immune evasion from the DBL crystal structure". Molecular and Biochemical Parasitology 148 (1): 1–9. July 2006. doi:10.1016/j.molbiopara.2006.03.004. PMID 16621067.
- ↑ "Complement receptor 1 and malaria". Cellular Microbiology 13 (10): 1441–50. October 2011. doi:10.1111/j.1462-5822.2011.01648.x. PMID 21790941.
- ↑ "DC8 and DC13 var genes associated with severe malaria bind avidly to diverse endothelial cells". PLOS Pathogens 9 (6): e1003430. 2013. doi:10.1371/journal.ppat.1003430. PMID 23825944.
- ↑ "Interaction between Endothelial Protein C Receptor and Intercellular Adhesion Molecule 1 to Mediate Binding of Plasmodium falciparum-Infected Erythrocytes to Endothelial Cells". mBio 7 (4): e00615–16. July 2016. doi:10.1128/mBio.00615-16. PMID 27406562.
- ↑ "Structural conservation despite huge sequence diversity allows EPCR binding by the PfEMP1 family implicated in severe childhood malaria". Cell Host & Microbe 17 (1): 118–29. January 2015. doi:10.1016/j.chom.2014.11.007. PMID 25482433.
- ↑ 72.0 72.1 "Full-length recombinant Plasmodium falciparum VAR2CSA binds specifically to CSPG and induces potent parasite adhesion-blocking antibodies". Journal of Molecular Biology 397 (3): 826–34. April 2010. doi:10.1016/j.jmb.2010.01.040. PMID 20109466.
- ↑ "Molecular basis of severe malaria". Proceedings of the National Academy of Sciences of the United States of America 109 (26): 10130–1. June 2012. doi:10.1073/pnas.1207174109. PMID 22679282. Bibcode: 2012PNAS..10910130D.
- ↑ Deshmukh, A. S.; Srivastava, S.; Dhar, S. K. (2013). "Plasmodium falciparum: Epigenetic Control of var Gene Regulation and Disease". in Kundu, T. K.. Epigenetics: Development and Disease. Subcellular Biochemistry. 61. Dordrecht: Springer. pp. 659–682. doi:10.1007/978-94-007-4525-4_28. ISBN 978-94-007-4524-7.
- ↑ "A restricted subset of var genes mediates adherence of Plasmodium falciparum-infected erythrocytes to brain endothelial cells". Proceedings of the National Academy of Sciences of the United States of America 109 (26): E1782-90. June 2012. doi:10.1073/pnas.1120534109. PMID 22619321.
- ↑ "A subset of group A-like var genes encodes the malaria parasite ligands for binding to human brain endothelial cells". Proceedings of the National Academy of Sciences of the United States of America 109 (26): E1772-81. June 2012. doi:10.1073/pnas.1120461109. PMID 22619330.
- ↑ "EPCR and Malaria Severity: The Center of a Perfect Storm". Trends in Parasitology 33 (4): 295–308. April 2017. doi:10.1016/j.pt.2016.11.004. PMID 27939609.
- ↑ "Plasmodium falciparum var genes expressed in children with severe malaria encode CIDRα1 domains". EMBO Molecular Medicine 8 (8): 839–50. August 2016. doi:10.15252/emmm.201606188. PMID 27354391.
- ↑ "var Genes Encoding Endothelial Protein C Receptor-Binding P. falciparum Erythrocyte Membrane Protein 1". Infection and Immunity 85 (4): IAI.00841–16. April 2017. doi:10.1128/IAI.00841-16. PMID 28138022.
- ↑ "Designing a VAR2CSA-based vaccine to prevent placental malaria". Vaccine 33 (52): 7483–8. December 2015. doi:10.1016/j.vaccine.2015.10.011. PMID 26469717.
 |