Biology:Plasmodium knowlesi
| Plasmodium knowlesi | |
|---|---|
| |
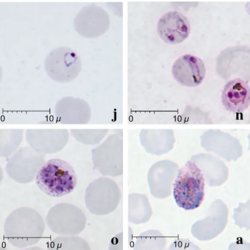
| Giemsa-stained smears of Plasmodium knowlesi infecting human red blood cells | |
| Scientific classification | |
| Domain: | Eukaryota |
| Clade: | Diaphoretickes |
| Clade: | SAR |
| Clade: | Alveolata |
| Phylum: | Apicomplexa |
| Class: | Aconoidasida |
| Order: | Haemospororida |
| Family: | Plasmodiidae |
| Genus: | Plasmodium |
| Species: | P. knowlesi
|
| Binomial name | |
| Plasmodium knowlesi Sinton and Mulligan 1932
| |
Plasmodium knowlesi is a parasite that causes malaria in humans and other primates. It is found throughout Southeast Asia, and is the most common cause of human malaria in Malaysia. Like other Plasmodium species, P. knowlesi has a life cycle that requires infection of both a mosquito and a warm-blooded host. While the natural warm-blooded hosts of P. knowlesi are likely various Old World monkeys, humans can be infected by P. knowlesi if they are fed upon by infected mosquitoes. P. knowlesi is a eukaryote in the phylum Apicomplexa, genus Plasmodium, and subgenus Plasmodium. It is most closely related to the human parasite Plasmodium vivax as well as other Plasmodium species that infect non-human primates.
Humans infected with P. knowlesi can develop uncomplicated or severe malaria similar to that caused by Plasmodium falciparum. Diagnosis of P. knowlesi infection is challenging as P. knowlesi very closely resembles other species that infect humans. Treatment is similar to other types of malaria, with chloroquine or artemisinin combination therapy typically recommended. P. knowlesi malaria is an emerging disease previously thought to be rare in humans, but increasingly recognized as a major health burden in Southeast Asia.
P. knowlesi was first described as a distinct species and as a potential cause of human malaria in 1932. It was briefly used in the early 20th century to cause fever as a treatment for neurosyphilis. In the mid-20th century, P. knowlesi became popular as a tool for studying Plasmodium biology and was used for basic research, vaccine research, and drug development. P. knowlesi is still used as a laboratory model for malaria, as it readily infects the model primate the rhesus macaque, and can be grown in cell culture in human or macaque blood.
Life cycle
File:Plasmodium life cycle CDC.tif Like other Plasmodium parasites, P. knowlesi has a life cycle that requires it be passed back and forth between mammalian hosts and insect hosts. Primates are infected through the bite of an infected Anopheles mosquito which carries a parasite stage called the sporozoite in its salivary glands. Sporozoites follow the blood stream to the primate liver where they develop and replicate over five to six days before bursting, releasing thousands of daughter cells called merozoites into the blood (unlike the related P. vivax, P. knowlesi does not make latent hypnozoites in the liver).[1][2] The merozoites in the blood attach to and invade the primate's red blood cells. Inside the red blood cell, the parasite progresses through several morphologically distinguishable stages, called the ring stage, the trophozoite, and the schizont. The schizont-infected red blood cells eventually burst, releasing up to 16 new merozoites into the blood stream that infect new red blood cells and continue the cycle.[1][3] P. knowlesi completes this red blood cell cycle every 24 hours, making it uniquely rapid among primate-infecting Plasmodium species (which generally take 48 or 72 hours).[2] Occasionally, parasites that invade red blood cells instead enter a sexual cycle, developing over approximately 48 hours into distinct sexual forms called microgametocytes or macrogametocytes.[1][3] These gametocytes remain in the blood to be ingested by mosquitoes.[1][3]
A mosquito ingests gametocytes when it takes a blood meal from an infected primate host. Once inside the mosquito gut, the gametocytes develop into gametes and then fuse to form a diploid zygote.[1] The zygote matures into an ookinete, which migrates through the wall of the mosquito gut and develops into an oocyst.[1][4] The oocyst then releases thousands of sporozoites, which migrate through the mosquito to the salivary glands.[1] This entire process in the mosquito takes 12 to 15 days.[2]
Cell biology

P. knowlesi largely resembles other Plasmodium species in its cell biology. Its genome consists of 23.5 megabases of DNA separated into 14 chromosomes.[5] It contains approximately 5200 protein-coding genes, 80% of which have orthologs present in P. falciparum and P. vivax.[5] The genome contains two large gene families that are unique to P. knowlesi: the SICAvar (schizont-infected cell agglutination variant) family, which is involved in displaying different antigens on the parasite surface to evade the immune system, and the Kir (knowlesi interspersed repeat) family, involved in adhering parasitized red blood cells to blood vessel walls.[5]
As an apicomplexan, P. knowlesi has several distinctive structures at its apical end that are specialized for invading host cells. These include the large bulbous rhoptries, smaller micronemes, and dispersed dense granules, each of which secretes effectors to enter and modify the host cell.[6][7] Like other apicomplexans, P. knowlesi also has two organelles of endosymbiotic origin: a single large mitochondrion and the apicoplast, both of which are involved in the parasite's metabolism.[8]
Evolution and taxonomy
Despite its morphological similarity to P. malariae, P. knowlesi is most closely related to P. vivax as well as other Plasmodium species that infect non-human primates.[9] The last common ancestor of all modern P. knowlesi strains lived an estimated 98,000 to 478,000 years ago.[9] Among human parasites, P. knowlesi is most closely related to P. vivax, from which it diverged between 18 million and 34 million years ago.[5][10] A phylogenetic tree comparing the Plasmodium species that infect humans is shown below:[5]
| Subgenus Plasmodium |
| ||||||||||||||||||
The population of P. knowlesi parasites is more genetically diverse than that of P. falciparum or P. vivax. Within P. knowlesi there are three genetically distinct subpopulations.[5] Two are present in the same areas of Malaysian Borneo and may infect different mosquitoes.[5] The third has been found only in laboratory isolates originating from other parts of Southeast Asia.[5] Populations of P. knowlesi isolated from macaques are genetically indistinguishable from those isolated from human infections, suggesting the same parasite populations can infect humans and macaques interchangeably.[11]
Three subspecies of P. knowlesi have been described based on differences in their appearance in stained blood films: P. knowlesi edesoni, P. knowlesi sintoni, and P. knowlesi arimai, which were isolated from Malaysia, Java, and Taiwan respectively.[2][12] The relationship between these described subspecies and the populations described in the modern literature is not clear.[2]
Distribution
Plasmodium knowlesi is found throughout Southeast Asia, where it primarily infects the long-tailed macaque, pig-tailed macaque, and Sumatran surili as well as the mosquito vectors Anopheles hackeri in peninsular Malaysia and Anopheles latens in Sarawak.[1] Long-tailed macaques in the wild can be infected with P. knowlesi [13] without any apparent disease, even when they are simultaneously infected with various other Plasmodium species.[2][10] P. knowlesi is rarely found outside of Southeast Asia, likely because the mosquitoes it infects are restricted to that region.[11]
Role in human disease

P. knowlesi can cause both uncomplicated and severe malaria in humans. Those infected nearly always experience fever and chills.[14] People with uncomplicated P. knowlesi malaria often also experience headaches, joint pain, malaise, and loss of appetite.[14] Less commonly, people report coughing, abdominal pain, diarrhea, nausea, and vomiting.[14] Laboratory tests of infected people nearly always show a low platelet count, although this rarely leads to bleeding problems.[14] Unlike other human malarias, P. knowlesi malaria tends to have fevers that spike every 24 hours, and is therefore often called daily or "quotidian" malaria.[14][15] Uncomplicated P. knowlesi malaria can be treated with antimalarial drugs.[14]
At least 10% of people infected with P. knowlesi develop severe malaria.[11] Severe P. knowlesi malaria resembles severe malaria caused by P. falciparum. Those with severe disease may experience shortness of breath, abdominal pain, and vomiting.[14] As disease progresses, parasites replicate to very high levels in the blood likely causing acute kidney injury, jaundice, shock, and respiratory distress.[14][16] Metabolic acidosis is uncommon, but can occur in particularly severe cases.[16] Unlike P. falciparum malaria, severe P. knowlesi malaria rarely causes coma or severe anemia.[14][16] Approximately 1-2% of cases are fatal.[11]
Diagnosis
Malaria is traditionally diagnosed by examining Giemsa-stained blood films under a microscope; however, differentiating P. knowlesi from other Plasmodium species in this way is challenging due to their similar appearance.[11] P. knowlesi ring-stage parasites stained with Giemsa resemble P. falciparum ring stages, appearing as a circle with one or two dark dots of chromatin.[17] Older trophozoites appear more dispersed, forming a rectangular-shape spread across the host cell called a "band-form" that resembles the similar stage in P. malariae.[17] During this stage, dots sometimes appear across the host red blood cell, called "Sinton and Mulligans' stippling".[17] Schizonts appear, similarly to other Plasmodium species, as clusters of purple merozoites surrounding a central dark-colored pigment.[17]
Due to the morphological similarity among Plasmodium species, misdiagnosis of P. knowlesi infection as P. falciparum, P. malariae, or P. vivax is common.[16] While some rapid diagnostic tests can detect P. knowlesi, they tend to have poor sensitivity and specificity and are therefore not always reliable.[16][18] Detection of nucleic acid by PCR or real-time PCR is the most reliable method for detecting P. knowlesi, and differentiating it from other Plasmodium species infection. However, due to the relatively slow and expensive nature of PCR, this is not available in many endemic areas.[11] Loop-mediated isothermal amplification methods of P. knowlesi detection have also been developed, but are not yet widely used.[11]
Treatment
Because P. knowlesi takes only 24 hours to complete its erythrocytic cycle, it can rapidly result in very high levels of parasitemia with fatal consequences.[16] For those with uncomplicated malaria, the World Health Organization recommends treatment with artemisinin-based combination therapy (ACT) or chloroquine.[19] For those with severe malaria, the World Health Organization recommends administration of intravenous artesunate for at least 24 hours, followed by ACT treatment.[16] Additionally, early drug trials have suggested that combinations of chloroquine and primaquine, artesunate and mefloquine, artemether and lumefantrine, and chloroquine alone could be effective treatments for uncomplicated P. knowlesi malaria.[16] There is no evidence of P. knowlesi developing resistance to current antimalarials.[16]
Epidemiology
P. knowlesi is the most common cause of malaria in Malaysia,[5] and cases of P. knowlesi malaria have been reported in most countries of Southeast Asia as well as travelers from the region.[11]
Infection with P. knowlesi is associated with socioeconomic and lifestyle factors that bring people into the dense forests where the mosquito hosts are commonly found.[14] In particular, those who work in the forest or at its margin such as farmers, hunters, and loggers are at increased risk for infection.[14] Likely for this reason, males are infected more frequently than females, and adults are infected more frequently than children.[14]
Research
P. knowlesi has long been used as a research model for studying the interaction between parasite and host, and developing antimalarial vaccines and drugs.[10] Its utility as a research model is partly due to its ability to infect rhesus macaques, a common laboratory model primate. Rhesus macaques are highly susceptible to P. knowlesi and can be infected by mosquito bite, injection of sporozoites, or injection of blood-stage parasites.[2][10] Infected monkeys develop some hallmarks of human malaria including anemia and enlargement of the spleen and liver.[2] Infection is typically fatal if untreated, with the cause of death seemingly circulatory failure characterized by adhesion of infected red blood cells to the blood vessel walls.[2] Monkeys can be cured of infection by treatment with antimalarials; repeated infection followed by cure results in the monkeys developing some immunity to infection, a topic that has also been the subject of substantial research.[2]
P. knowlesi is also used for in vitro research into Plasmodium cell biology. Isolated sporozoites can infect primary rhesus hepatocytes, allowing the in vitro study of the parasite liver stage.[10] Additionally, P. knowlesi and P. falciparum are the only Plasmodium species that can be maintained continuously in cultured red blood cells, both rhesus and human.[10] Facilitating molecular biology research, the P. knowlesi genome has been sequenced and is available on PlasmoDB and other online repositories.[10] P. knowlesi can be genetically modified in the lab by transfection either in the rhesus macaque model system, or in blood cell culture.[10][20] Blood-infecting stages and sporozoites can be stored long-term by freezing with glycerolyte, allowing the preservation of strains of interest.[1]
History

The Italian physician Giuseppe Franchini first described what may have been P. knowlesi in 1927 when he noted a parasite distinct from P. cynomolgi and P. inui in the blood of a long-tailed macaque.[21][22] In 1931, the parasite was again seen in a long-tailed macaque by H. G. M. Campbell during his work on kala azar (visceral leishmaniasis) in Calcutta; Campbell's colleague Lionel Everard Napier drew blood from the affected monkey and inoculated three laboratory monkeys, one of which was a rhesus macaque that developed a severe infection.[3][23] Campbell and Napier gave the infected monkey to Biraj Mohan Das Gupta who was able to maintain the parasite by serial passage through monkeys.[24] In 1932, Das Gupta and his supervisor Robert Knowles described the morphology of the parasite in macaque blood, and demonstrated that it could infect three human patients (in each case it was used to induce fever with the hope of treating another infection).[3][25] Also in 1932, John Sinton and H. W. Mulligan further described the morphology of the parasite in blood cells, determined it to be a distinct species from others described, and named it Plasmodium knowlesi in honor of Robert Knowles.[3]
Soon thereafter, in 1935 C. E. Van Rooyen and George R. Pile reported using P. knowlesi infection to treat general paralysis in psychiatric patients. P. knowlesi would go on to be used as a general pyretic agent for various diseases, particularly neurosyphilis for which it was used until at least 1955.[24] While Cyril Garnham had suggested in 1957 that P. knowlesi might naturally infect humans,[26] the first documented case of a human naturally infected with P. knowlesi was in 1965 in a U.S. Army surveyor who developed chills and fever after a five-day deployment in Malaysia.[24][27] Based on this finding, a team at the Institute for Medical Research in Peninsular Malaysia undertook a survey of people living in proximity to macaques, but failed to find evidence that simian malaria was being transmitted to humans.[24]
Through the 1960s and 1970s, scientific research groups used P. knowlesi as a research model to make seminal discoveries in malaria. In 1965 and 1972, several groups characterized how P. knowlesi antigenic variation contributed to immune evasion and chronic infection.[24] In 1975, Louis H. Miller and others showed that P. knowlesi required Duffy factor on the surface of red blood cells in order to invade them (they would go on to show the same requirement for P. vivax a year later).[24]
Work on P. knowlesi as a human malaria parasite was revitalized in 2004, when Balbir Singh and others used PCR to show that over half of a group of humans diagnosed with P. malariae malaria in Malaysian Borneo were actually infected with P. knowlesi.[24][28] Over the following decade, several investigators used molecular detection methods capable of distinguishing P. knowlesi from morphologically similar parasites to attribute an increasing proportion of malaria cases to P. knowlesi throughout Southeast Asia.[29] Work with archival samples has shown that infection with this parasite has occurred in Malaysia at least since the 1990s.[30]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 "Plasmodium knowlesi: A Malaria Parasite of Monkeys and Humans". Annual Review of Entomology 57: 107–121. 2012. doi:10.1146/annurev-ento-121510-133540. PMID 22149265. https://zenodo.org/record/1234957.
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 "The role of Plasmodium knowlesi in the history of malaria research". Parasitology (Cambridge University Press) 145 (1): 6–17. September 2016. doi:10.1017/S0031182016001888. PMID 27829470.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 The Primate Malarias. Atlanta, GA: Division of Parasitic Diseases, U.S. Centers for Disease Control. 1971. pp. 317–334. https://stacks.cdc.gov/view/cdc/6538. Retrieved 26 August 2019.
- ↑ "Plasmodium Oocysts: Overlooked Targets of Mosquito Immunity". Trends in Parasitology (Elsevier) 32 (12): 979–990. December 2016. doi:10.1016/j.pt.2016.08.012. PMID 27639778. https://lib.dr.iastate.edu/ent_pubs/435.
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 5.7 5.8 "Plasmodium genomics: an approach for learning about and ending human malaria". Parasitology Research (Springer) 118 (1): 1–27. January 2019. doi:10.1007/s00436-018-6127-9. PMID 30402656.
- ↑ Counihan, Natalie A.; Kalanon, Ming; Coppel, Ross L.; De Koning-Ward, Tania F. (2013). "Plasmodium rhoptry proteins: Why order is important". Trends in Parasitology 29 (5): 228–36. doi:10.1016/j.pt.2013.03.003. PMID 23570755.
- ↑ Kemp, Louise E.; Yamamoto, Masahiro; Soldati-Favre, Dominique (2013). "Subversion of host cellular functions by the apicomplexan parasites". FEMS Microbiology Reviews 37 (4): 607–31. doi:10.1111/1574-6976.12013. PMID 23186105. https://archive-ouverte.unige.ch/unige:28792/ATTACHMENT01.
- ↑ Sheiner, Lilach; Vaidya, Akhil B.; McFadden, Geoffrey I. (2013). "The metabolic roles of the endosymbiotic organelles of Toxoplasma and Plasmodium spp". Current Opinion in Microbiology 16 (4): 452–8. doi:10.1016/j.mib.2013.07.003. PMID 23927894.
- ↑ 9.0 9.1 Kazura, James W., ed (2011). "Plasmodium knowlesi: Reservoir Hosts and Tracking the Emergence in Humans and Macaques". PLOS Pathog 7 (4): e1002015. doi:10.1371/journal.ppat.1002015. PMID 21490952.
- ↑ 10.0 10.1 10.2 10.3 10.4 10.5 10.6 10.7 "Plasmodium knowlesi: a relevant, versatile experimental malaria model". Parasitology (Cambridge University Press) 145 (1): 56–70. November 2016. doi:10.1017/S0031182016002286. PMID 27938428.
- ↑ 11.0 11.1 11.2 11.3 11.4 11.5 11.6 11.7 "Human infections with Plasmodium knowlesi-zoonotic malaria". Clinical Microbiology and Infection (Elsevier) 21 (7): 640–648. July 2015. doi:10.1016/j.cmi.2015.03.017. PMID 25843504.
- ↑ Garnham PC (1963). "A new sub-species of Plasmodium knowlesi in the long-tailed macaque.". J Trop Med Hyg 66: 156–8. PMID 13960457. https://pubmed.ncbi.nlm.nih.gov/13960457.
- ↑ Yusuf, Noorazian Md; Zulkefli, Jannah; Jiram, Adela Ida; Vythilingam, Indra; Hisam, Shamilah; Devi, Renuka; Salehhuddin, Afiqah; Ali, Nurulshuhada Md et al. (2022). "Plasmodium spp. in macaques, Macaca fascicularis, in Malaysia, and their potential role in zoonotic malaria transmission". Parasite (EDP Sciences) 29: 32. doi:10.1051/parasite/2022032. ISSN 1776-1042. PMID 35674419.
- ↑ 14.00 14.01 14.02 14.03 14.04 14.05 14.06 14.07 14.08 14.09 14.10 14.11 "Human Infections and Detection of Plasmodium knowlesi". Clinical Microbiology Reviews 26 (2): 165–184. 2013. doi:10.1128/CMR.00079-12. PMID 23554413.
- ↑ "A naturally acquired quotidian-type malaria in man transferable to monkeys". Science 149 (3686): 865. 1965. doi:10.1126/science.149.3686.865. PMID 14332847. Bibcode: 1965Sci...149..865C.
- ↑ 16.0 16.1 16.2 16.3 16.4 16.5 16.6 16.7 16.8 "The treatment of Plasmodium knowlesi malaria". Trends in Parasitology (Elsevier) 33 (3): 242–253. March 2017. doi:10.1016/j.pt.2016.09.002. PMID 27707609.
- ↑ 17.0 17.1 17.2 17.3 "DPDx Malaria - Image Gallery". U.S. Centers for Disease Control and Prevention. 29 December 2017. https://www.cdc.gov/dpdx/malaria/index.html.
- ↑ Parasitic Diseases (7 ed.). Parasites Without Borders. p. 112. https://s3.amazonaws.com/parasiteswithoutborders/PD+7th+Edition+Low+Res+Version+5-11-2019.pdf.
- ↑ "Treatment of uncomplicated malaria caused by P. vivax, P. ovale, P. malariae, or P. knowlesi". Guidelines for the Treatment of Malaria (3 ed.). World Health Organization. 2015. p. 60. ISBN 978-92-4-154912-7. https://www.who.int/malaria/publications/atoz/9789241549127/en/. Retrieved 22 January 2020.
- ↑ de Koning-Ward TF; Gilson PR; Crabb BS (June 2015). "Advances in molecular genetic systems in malaria". Nature Reviews Microbiology (Macmillan) 13 (6): 373–387. doi:10.1038/nrmicro3450. PMID 25978707.
- ↑ Franchini G (1927) Su di un plasmodio pigmentato di una scimmia. Arch Ital Sci Med Colon 8:187–90
- ↑ "Plasmodium knowlesi: an overlooked Italian discovery?". Clinical Infectious Diseases 53 (8): 849; author reply 849–50. October 2011. doi:10.1093/cid/cir527. PMID 21890752.
- ↑ Napier LE; Campbell HGM (May 1932). "Observations on a Plasmodium infection which causes haemoglobinuria in certain species of monkey". The Indian Medical Gazette: 246–249. http://pdfs.semanticscholar.org/8cad/758a4a40a3f5802e264e4abc059d9a9eb775.pdf. Retrieved 27 August 2019.
- ↑ 24.0 24.1 24.2 24.3 24.4 24.5 24.6 "Plasmodium knowlesi: The emerging zoonotic malaria parasite". Acta Tropica 125 (2): 191–201. 2013. doi:10.1016/j.actatropica.2012.10.008. PMID 23088834.
- ↑ "A study of monkey malaria, and its experimental transmission to man". Indian Medical Gazette 67 (6): 301–320. June 1932. PMID 29010910.
- ↑ "The tissue stages and sporogony of Plasmodium knowlesi". Trans R Soc Trop Med Hyg 51 (5): 384–396. 1957. doi:10.1016/0035-9203(57)90071-8. PMID 13467997.
- ↑ Mason Dentinger, R (26 August 2015). "Patterns of Infection and Patterns of Evolution: How a Malaria Parasite Brought "Monkeys and Man" Closer Together in the 1960s". Journal of the History of Biology 49 (2): 359–395. doi:10.1007/s10739-015-9421-8. PMID 26307748.
- ↑ "A large focus of naturally acquired Plasmodium knowlesi infections in human beings". Lancet 363 (9414): 1017–24. 2004. doi:10.1016/S0140-6736(04)15836-4. PMID 15051281. http://ir.unimas.my/2202/1/A%20large%20focus%20of%20naturally%20acquired%20Plasmodium%20knowlesi%20infections%20in%20human%20beings.pdf.
- ↑ "Plasmodium knowlesi in humans, macaques and mosquitoes in peninsular Malaysia". Parasit Vectors 1 (1): 26. 2008. doi:10.1186/1756-3305-1-26. PMID 18710577.
- ↑ Lee KS; Cox-Singh J; Brooke G; Matusop A; Singh B (2009). "Plasmodium knowlesi from archival blood films: Further evidence that human infections are widely distributed and not newly emergent in Malaysian Borneo". Int J Parasitol 39 (10): 1125–1128. doi:10.1016/j.ijpara.2009.03.003. PMID 19358848.
External links
- CDC malaria page
- WHO malaria page
- P. knowlesi genome data
- Short film on the discovery of the role of P. knowlesi in human malaria
Wikidata ☰ Q241594 entry
 |
