Biology:Cro repressor family
| Cro | |||||||||
|---|---|---|---|---|---|---|---|---|---|
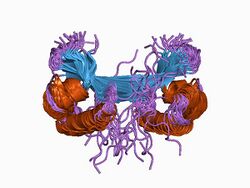 three-dimensional dimer structure of the lambda-cro repressor in solution as determined by heteronuclear multidimensional nmr | |||||||||
| Identifiers | |||||||||
| Symbol | Cro | ||||||||
| Pfam | PF09048 | ||||||||
| Pfam clan | CL0123 | ||||||||
| InterPro | IPR000655 | ||||||||
| |||||||||
In molecular biology, the Cro repressor family is a family of repressor proteins in bacteriophage lambda that includes the Cro repressor.
Bacteriophage lambda encodes two repressors: the Cro repressor that acts to turn off early gene transcription during the lytic cycle, and the lambda or cI repressor required to maintain lysogenic growth. Together the Cro and cI repressors form a helix-turn-helix (HTH) superfamily. The lambda Cro repressor binds to DNA as a highly flexible dimer. The crystal structure of the lambda Cro repressor reveals a HTH DNA-binding protein with an alpha/beta fold that differs from other Cro family members, possibly by an evolutionary fold change.[1][2] Most Cro proteins, such as Enterobacteria phage P22 Cro and Bacteriophage 434 Cro, have an all-alpha structure that is thought to be ancestral to lambda Cro, where the fourth and fifth helices are replaced by a beta-sheet, possibly as a result of secondary structure switching rather than by nonhomologous replacement.[3]
References
- ↑ "Refined structure of Cro repressor protein from bacteriophage lambda suggests both flexibility and plasticity". J. Mol. Biol. 280 (1): 129–36. July 1998. doi:10.1006/jmbi.1998.1849. PMID 9653036.
- ↑ "Reduced immune adherence of antigen/antibody complexes formed in the presence of complement in vivo and in vitro". Complement Inflamm 6 (6): 470–9. 1989. doi:10.1159/000463116. PMID 2598646.
- ↑ "Secondary structure switching in Cro protein evolution". Structure 12 (4): 569–81. April 2004. doi:10.1016/j.str.2004.02.024. PMID 15062080.
 |

