Biology:ATP-binding domain of ABC transporters
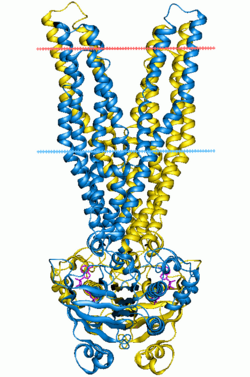 Multidrug ABC transporter SAV1866, closed state | |||||||||||
| Identifiers | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Symbol | ABC_tran | ||||||||||
| Pfam | PF00005 | ||||||||||
| InterPro | IPR003439 | ||||||||||
| PROSITE | PDOC00185 | ||||||||||
| SCOP2 | 1b0u / SCOPe / SUPFAM | ||||||||||
| TCDB | 3.A.1 | ||||||||||
| OPM superfamily | 17 | ||||||||||
| OPM protein | 2hyd | ||||||||||
| CDD | cd00267 | ||||||||||
| |||||||||||
In molecular biology, ATP-binding domain of ABC transporters is a water-soluble domain of transmembrane ABC transporters.
ABC transporters belong to the ATP-Binding Cassette superfamily, which uses the hydrolysis of ATP to translocate a variety of compounds across biological membranes. ABC transporters are minimally constituted of two conserved regions: a highly conserved ATP binding cassette (ABC) and a less conserved transmembrane domain (TMD). These regions can be found on the same protein or on two different ones. Most ABC transporters function as a dimer and therefore are constituted of four domains, two ABC modules and two TMDs.
Biological function
ABC transporters are involved in the export or import of a wide variety of substrates ranging from small ions to macromolecules. The major function of ABC import systems is to provide essential nutrients to bacteria. They are found only in prokaryotes and their four constitutive domains are usually encoded by independent polypeptides (two ABC proteins and two TMD proteins). Prokaryotic importers require additional extracytoplasmic binding proteins (one or more per systems) for function. In contrast, export systems are involved in the extrusion of noxious substances, the export of extracellular toxins and the targeting of membrane components. They are found in all living organisms and in general the TMD is fused to the ABC module in a variety of combinations. Some eukaryotic exporters encode the four domains on the same polypeptide chain.
Amino acid sequence
The ABC module (approximately two hundred amino acid residues) is known to bind and hydrolyze ATP, thereby coupling transport to ATP hydrolysis in a large number of biological processes. The cassette is duplicated in several subfamilies. Its primary sequence is highly conserved, displaying a typical phosphate-binding loop: Walker A, and a magnesium binding site: Walker B. Besides these two regions, three other conserved motifs are present in the ABC cassette: the switch region which contains a histidine loop, postulated to polarize the attacking water molecule for hydrolysis, the signature conserved motif (LSGGQ) specific to the ABC transporter, and the Q-motif (between Walker A and the signature), which interacts with the gamma phosphate through a water bond. The Walker A, Walker B, Q-loop and switch region form the nucleotide binding site.
3D structure
The 3D structure of a monomeric ABC module adopts a stubby L-shape with two distinct arms.[1][2] ArmI (mainly beta-strand) contains Walker A and Walker B. The important residues for ATP hydrolysis and/or binding are located in the P-loop. The ATP-binding pocket is located at the extremity of armI. The perpendicular armII contains mostly the alpha helical subdomain with the signature motif. It only seems to be required for structural integrity of the ABC module. ArmII is in direct contact with the TMD. The hinge between armI and armII contains both the histidine loop and the Q-loop, making contact with the gamma phosphate of the ATP molecule. ATP hydrolysis leads to a conformational change that could facilitate ADP release. In the dimer the two ABC cassettes contact each other through hydrophobic interactions at the antiparallel beta-sheet of armI by a two-fold axis.
Human proteins containing this domain
ABCA1; ABCA10; ABCA12; ABCA13; ABCA2; ABCA3; ABCA4; ABCA5; ABCA6; ABCA7; ABCA8; ABCA9; ABCB1; ABCB10; ABCB11; ABCB4; ABCB5; ABCB6; ABCB7; ABCB8; ABCB9; ABCC1; ABCC10; ABCC11; ABCC12; ABCC2; ABCC3; ABCC4; ABCC5; ABCC6; ABCC8; ABCC9; ABCD1; ABCD2; ABCD3; ABCD4; ABCE1; ABCF1; ABCF2; ABCF3; ABCG1; ABCG2; ABCG4; ABCG5; ABCG8; CFTR; TAP1; TAP2; TAPL;
References
- ↑ "Crystal structure of the ATP-binding subunit of an ABC transporter". Nature 396 (6712): 703–7. December 1998. doi:10.1038/25393. PMID 9872322.
- ↑ "Structure and mechanism of ABC transporter proteins". Curr. Opin. Struct. Biol. 17 (4): 412–8. August 2007. doi:10.1016/j.sbi.2007.07.003. PMID 17723295.
- Rosteck Jr, P. R.; Reynolds, P. A.; Hershberger, C. L. (1991). "Homology between proteins controlling Streptomyces fradiae tylosin resistance and ATP-binding transport". Gene 102 (1): 27–32. doi:10.1016/0378-1119(91)90533-h. PMID 1864505.
- Blight, M. A.; Holland, I. B. (1990). "Structure and function of haemolysin B,P-glycoprotein and other members of a novel family of membrane translocators". Molecular Microbiology 4 (6): 873–880. doi:10.1111/j.1365-2958.1990.tb00660.x. PMID 1977073.
- Higgins, C. F.; Hyde, S. C.; Mimmack, M. M.; Gileadi, U.; Gill, D. R.; Gallagher, M. P. (1990). "Binding protein-dependent transport systems". Journal of Bioenergetics and Biomembranes 22 (4): 571–592. doi:10.1007/BF00762962. PMID 2229036.
 |

