Biology:Actomyosin ring
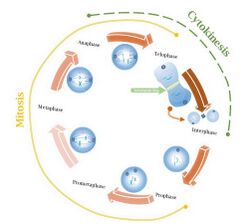
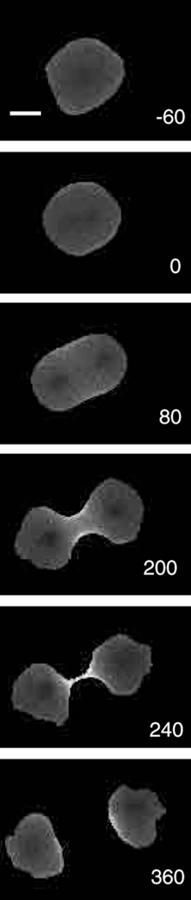
In molecular biology, an actomyosin ring or contractile ring, is a prominent structure during cytokinesis.[1] It forms perpendicular to the axis of the spindle apparatus[2] towards the end of telophase, in which sister chromatids are identically separated at the opposite sides of the spindle forming nuclei (Figure 1). The actomyosin ring follows an orderly sequence of events: identification of the active division site, formation of the ring, constriction of the ring, and disassembly of the ring.[1] It is composed of actin and myosin II bundles, thus the term actomyosin. The actomyosin ring operates in contractile motion,[3] although the mechanism on how or what triggers the constriction is still an evolving topic.[4] Other cytoskeletal proteins are also involved in maintaining the stability of the ring[5] and driving its constriction.[6] Apart from cytokinesis, in which the ring constricts as the cells divide (Figure 2), actomyosin ring constriction has also been found to activate during wound closure.[7] During this process, actin filaments are degraded, preserving the thickness of the ring. After cytokinesis is complete, one of the two daughter cells inherits a remnant known as the midbody ring.[8]
Activation of the cell-cycle kinase (e.g. Rho-kinases) during telophase initiates constriction of the actomyosin ring by creating a groove that migrates in an inward motion. Rho-kinases such as ROCK1 has been found to regulate actomyosin contraction through phosphorylation of the myosin light chain (MLC).[9] This mechanism promotes cell-cell contacts and integrity leading to adhesion formation.
Variation between kingdoms
In animals, the ring forms along the cleavage furrow on the inside of the plasma membrane then splits by abscission.[10][11] In fungi, it forms at the mother-bud neck before mitosis. Septin is heavily involved in the formation of the fungal AMR.[12] In most bacteria and many archaea a homologous structure called the Z-ring forms out of FtsZ, a homolog of tubulin.[13] Chloroplasts form an analogous structure out of FtsZ. These structures are not made out of actomyosin, but serve a similar role in constricting and permitting cytokinesis. In plant cells, there is no actomyosin ring. Instead, a cell plate grows centrifugally outwards from the center of the plane of division until it fuses with the existing cell wall.[14]
See also
References
- ↑ 1.0 1.1 Cheffings, T. H.; Burroughs, N. J.; Balasubramanian, M. K. (2016). "Actomyosin Ring Formation and Tension Generation in Eukaryotic Cytokinesis". Current Biology 26 (15): R719–R737. doi:10.1016/j.cub.2016.06.071. PMID 27505246.
- ↑ Mana-Capelli, Sebastian; McCollum, Dannel (2013). "Actomyosin Ring". Encyclopedia of Systems Biology. p. 8. doi:10.1007/978-1-4419-9863-7_779. ISBN 978-1-4419-9862-0.
- ↑ Munjal, A.; Lecuit, T. (2014). "Actomyosin networks and tissue morphogenesis". Development 141 (9): 1789–1793. doi:10.1242/dev.091645. PMID 24757001.
- ↑ Mendes Pinto, I.; Rubinstein, B.; Li, R. (2013). "Force to divide: Structural and mechanical requirements for actomyosin ring contraction". Biophysical Journal 105 (3): 547–554. doi:10.1016/j.bpj.2013.06.033. PMID 23931302.
- ↑ Martin, A. C. (2016). "Embryonic ring closure: Actomyosin rings do the two-step". The Journal of Cell Biology 215 (3): 301–303. doi:10.1083/jcb.201610061. PMID 27799371.
- ↑ Kucera, Ondrej; Siahaan, Valerie; Janda, Daniel; Dijkstra, Sietske H; Pilatova, Eliska; Zatecka, Eva; Diez, Stefan; Braun, Marcus et al. (2021). "Anillin propels myosin-independent constriction of actin rings". Nature Communications 12 (1): 4595. doi:10.1038/s41467-021-24474-1. PMID 34321459.
- ↑ Schwayer, C.; Sikora, M.; Slováková, J.; Kardos, R.; Heisenberg, C. P. (2016). "Actin Rings of Power". Developmental Cell 37 (6): 493–506. doi:10.1016/j.devcel.2016.05.024. PMID 27326928.
- ↑ Pelletier, L.; Yamashita, Y. M. (2012). "Centrosome asymmetry and inheritance during animal development". Current Opinion in Cell Biology 24 (4): 541–546. doi:10.1016/j.ceb.2012.05.005. PMID 22683192.
- ↑ Shi, J.; Surma, M.; Zhang, L.; Wei, L. (2013). "Dissecting the roles of ROCK isoforms in stress-induced cell detachment". Cell Cycle 12 (10): 1492–1900. doi:10.4161/cc.24699. PMID 23598717.
- ↑ Chen, C. T.; Hehnly, H.; Doxsey, S. J. (2012). "Orchestrating vesicle transport, ESCRTs and kinase surveillance during abscission". Nature Reviews. Molecular Cell Biology 13 (8): 483–488. doi:10.1038/nrm3395. PMID 22781903.
- ↑ Fededa, J. P.; Gerlich, D. W. (2012). "Molecular control of animal cell cytokinesis". Nature Cell Biology 14 (5): 440–447. doi:10.1038/ncb2482. PMID 22552143.
- ↑ Meitinger, F.; Palani, S. (2016). "Actomyosin ring driven cytokinesis in budding yeast". Seminars in Cell & Developmental Biology 53: 19–27. doi:10.1016/j.semcdb.2016.01.043. PMID 26845196.
- ↑ Alberts, B., A. Johnson, J. Lewis, D. Morgan, M. Raff, K. Roberts, and P. Walter, editors. (2015). Molecular Biology of the Cell, 6th edition. Garland Science: New York. 1464 pp.
- ↑ Bi, E.; Maddox, P.; Lew, D. J.; Salmon, E. D.; McMillan, J. N.; Yeh, E.; Pringle, J. R. (1998). "Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis". The Journal of Cell Biology 142 (5): 1301–1312. doi:10.1083/jcb.142.5.1301. PMID 9732290.
 |