Biology:Azurin
| Azurin | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Azurin tetramer, Pseudomonas aeruginosa | |||||||||
| Identifiers | |||||||||
| Symbol | Copper-bind | ||||||||
| Pfam | PF00127 | ||||||||
| SCOP2 | 5azu / SCOPe / SUPFAM | ||||||||
| CDD | cd13922 | ||||||||
| |||||||||
Azurin is a small, periplasmic, bacterial blue copper protein found in Pseudomonas, Bordetella, or Alcaligenes bacteria. Azurin moderates single-electron transfer between enzymes associated with the cytochrome chain by undergoing oxidation-reduction between Cu(I) and Cu(II). Each monomer of an azurin tetramer has a molecular weight of approximately 14kDa, contains a single copper atom, is intensively blue, and has a fluorescence emission band centered at 308 nm.
Azurins and pseudoazurins participate in the denitrification processes in bacteria.,[1] including the gram-negative bacteria Pseudomonas aeruginosa, by interacting with cytochrome c551. Azurin from P aeruginosa is a type I blue copper protein (cupredoxin), while cytochrome c551 (9 kDa) is a haem-containing cytochrome. Azurin possesses a relatively large hydrophobic patch close to the active site, and two residues in this hydrophobic patch, Met-44 and Met-64, are believed to be involved in its interaction with the redox partners cytochrome c551 and nitrite reductase.[2]
Although unrelated to its electron-transfer property, azurin has been found to have anticancer properties through its interaction with tumor-suppressor protein p53.
Protein function
In its oxidized form, azurin (Cu2+Az) receives an electron from its redox partner and is reduced according to the following reaction:
- Cu2+Az + e− → Cu+Az
The redox potential is 310 mV.[3]
The highly-interconnected beta-sheet structure of azurin is strongly coupled with its electron-transfer center (the copper-binding side).[4] Considerable experimental evidence exists to suggest that hydrogen bonds play a role in the long-distance electron transfer mechanism of azurin. Taken together, these observations suggest that electrons tunnel through the protein along its polypeptide and hydrogen bonds, making azurin a useful model system for studying long-range, intraprotein electron transfer (LRET).[4]
Protein structure
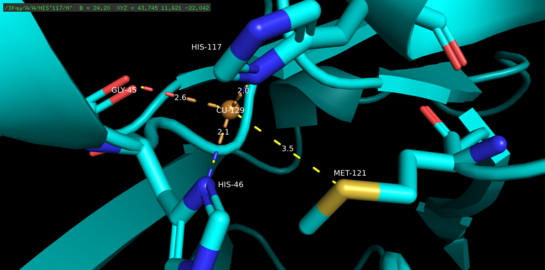
Azurin is a monomeric protein that weighs approximately 14 kDa and is composed of 128 amino acids forming eight beta-strands arranged in a beta-barrel formation.[5] The strands are connected by turns and a single alpha-helical insertion.[5] A single-atom copper binding site is located about 7 Å below each monomer’s surface towards its northern end; the copper atom that inhabits it is coordinated by five ligands surrounded by an extensive hydrophobic patch.[6]
The three equatorial copper ligands are composed of a thiolate (Cys112) and two imidazoles (His46, His117), and the carbonyl oxygen atoms of Gly45 and Met121 serve as the two weak axial ligands.[5] With the exception of Gly45, the copper-binding configuration above is common to the structures of all blue type 1 copper-binding proteins determined thus far.[7] Once coordinated, the ligand-metal complex assumes a distorted, trigonal bi-pyramidal geometry that stabilizes the cuprous (Cu(I)) reduced state of the protein relative to the cupric (Cu(II)) oxidized state.[5] Structurally imposed backbonding between the copper d orbitals and its ligand p orbitals may further stabilize the cuprous state.[8]
Existing structural information about azurin has largely been derived from X-ray crystallography studies of single-site mutated forms of the protein. Notable structural features elucidated by crystallography include the beta-sandwich motif formed from eight interlocking beta strands,[5] as well as an alpha-helical segment outside the barrel linking beta-sheets 4 and 5.[5]
Although the Cu(I)/Cu(II) redox potential is typically higher for azurin than most other copper complexes, structural studies in which Met121 (one of azurin’s equatorial copper-coordinating ligands) is replaced have demonstrated that the absence of a thiolate copper ligand does not preclude high reduction potentials, as large hydrophobic residues in position 121 also raise the redox potential of the copper atom.[8] Thus, the higher redox potentials have been attributed to the exclusion of water from the metal-binding site, a condition augmented by the presence of bulky hydrophobic residues.[8]
Conversely, negatively charged residues lower the redox potential, since they stabilize the more positively charged cupric form of the copper ion.[8]
Biological function
When expressed in nitrogen-fixing organisms, azurin serves as the electron donor to nitrite reductase, an enzyme in the denitrification pathway of the nitrogen cycle.[9]
Azurins support oxidative deamination of primary amines by passing electrons from aromatic amine dehydrogenase to cytochrome oxidase, as well as from some c-type cytochromes to nitrite reductases.[10]
Disease relevance
Azurin has garnered significant attention as a potential therapeutic for various diseases, including cancer.[11] In vivo, it has been demonstrated to induce regression of human melanoma and breast cancer tissue with minimal toxic effects to the organism.[11]
Azurin enters preferentially into cancer cells via the p28 domain of the enzyme, which roughly corresponds to the extended alpha-helical region of the enzyme.[11] In cancer cells, azurin complexes with p53, stabilizing it and preventing association with E3 ubiquitin ligases, which would otherwise bind and mark the protein for destruction.[12] Four azurin molecules bind each p53 monomer with high affinity.[12] The p53/azurin complex travels to the nucleus, where p53 upregulates the transcription of proapoptotic genes Bax and Noxa.[12] P53 also activates the expression of cell-cycle inhibitors, preventing tumor cells from progressing beyond the G1 or S phase.[12] Although this pathway plays a significant role in azurin’s anticancer activity, the details of the interaction between azurin and p53 are not well understood.
A phase I clinical trial in the United States demonstrated both partial and complete tumor regression effects in fifteen stage IV cancer patients treated with the p28 amino-acid fragment of azurin.[13] Another phase I trial with the p28 fragment demonstrated azurin’s therapeutic effects against pediatric patients with brain tumors; subsequently, the USFDA approved the designation of p28 as an orphan drug for glioma.[14]
Azurin’s other domains may also exhibit strong anticancer activity by binding to cell surface receptor tyrosine kinases such as EphB2 receptors, which induce angiogenesis in cancer cells.[14] This is another mechanism by which azurin has been proposed to exhibit its therapeutic effects.
See also
References
- ↑ "Blue copper proteins: a comparative analysis of their molecular interaction properties". Protein Science 9 (8): 1439–54. August 2000. doi:10.1110/ps.9.8.1439. PMID 10975566.
- ↑ "The bacterial redox protein azurin induces apoptosis in J774 macrophages through complex formation and stabilization of the tumor suppressor protein p53". Infection and Immunity 70 (12): 7054–62. December 2002. doi:10.1128/IAI.70.12.7054-7062.2002. PMID 12438386.
- ↑ "Calculation of the redox potential of the protein azurin and some mutants". ChemBioChem 6 (4): 738–46. April 2005. doi:10.1002/cbic.200400244. PMID 15747387.
- ↑ 4.0 4.1 "Long-term molecular dynamics simulation of copper azurin: structure, dynamics and functionality". Biophysical Chemistry 78 (3): 247–57. April 1999. doi:10.1016/S0301-4622(99)00029-0. PMID 17030312.
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 "Crystal structure determinations of oxidized and reduced pseudoazurins from Achromobacter cycloclastes. Concerted movement of copper site in redox forms with the rearrangement of hydrogen bond at a remote histidine". The Journal of Biological Chemistry 274 (25): 17845–52. June 1999. doi:10.1074/jbc.274.25.17845. PMID 10364229.
- ↑ "Involvement of the hydrophobic patch of azurin in the electron-transfer reactions with cytochrome C551 and nitrite reductase". European Journal of Biochemistry 194 (1): 109–18. November 1990. doi:10.1111/j.1432-1033.1990.tb19434.x. PMID 2174771.
- ↑ "Structural biology of metal-binding sequences". Current Opinion in Chemical Biology 6 (2): 217–23. April 2002. doi:10.1016/S1367-5931(02)00314-9. PMID 12039007.
- ↑ 8.0 8.1 8.2 8.3 "Azurin as a protein scaffold for a low-coordinate nonheme iron site with a small-molecule binding pocket". Journal of the American Chemical Society 134 (48): 19746–57. December 2012. doi:10.1021/ja308346b. PMID 23167247.
- ↑ "In vivo studies disprove an obligatory role of azurin in denitrification in Pseudomonas aeruginosa and show that azu expression is under control of rpoS and ANR". Microbiology 143 (9): 2853–63. September 1997. doi:10.1099/00221287-143-9-2853. PMID 9308169.
- ↑ "Evidence for two distinct azurins in Alcaligenes xylosoxidans (NCIMB 11015): potential electron donors to nitrite reductase". Biochemistry 34 (32): 10180–6. August 1995. doi:10.1021/bi00032a011. PMID 7640272.
- ↑ 11.0 11.1 11.2 "Bacterial protein azurin as a new candidate drug to treat untreatable breast cancers". 1st Portuguese Biomedical Engineering Meeting. March 2011. pp. 1–4. doi:10.1109/ENBENG.2011.6026047. ISBN 978-1-4577-0522-9.
- ↑ 12.0 12.1 12.2 12.3 "Bacterial cupredoxin azurin hijacks cellular signaling networks: Protein-protein interactions and cancer therapy". Protein Science 26 (12): 2334–2341. December 2017. doi:10.1002/pro.3310. PMID 28960574.
- ↑ "A first-in-class, first-in-human, phase I trial of p28, a non-HDM2-mediated peptide inhibitor of p53 ubiquitination in patients with advanced solid tumours". British Journal of Cancer 108 (5): 1061–70. March 2013. doi:10.1038/bjc.2013.74. PMID 23449360.
- ↑ 14.0 14.1 "Cupredoxin-cancer interrelationship: azurin binding with EphB2, interference in EphB2 tyrosine phosphorylation, and inhibition of cancer growth". Biochemistry 46 (7): 1799–810. February 2007. doi:10.1021/bi061661x. PMID 17249693.
 |
