Biology:Bacillaceae-1 RNA motif
From HandWiki
| Bacillaceae-1 RNA | |
|---|---|
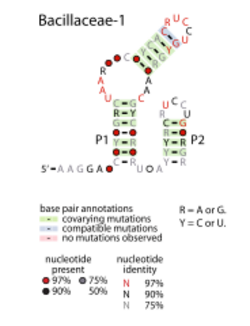 Consensus secondary structure of Bacilliceae-1 RNAs | |
| Identifiers | |
| Symbol | Bacillaceae-1 |
| Rfam | RF01690 |
| Other data | |
| Domain(s) | bacillaceae |
| PDB structures | PDBe |
The Bacillaceae-1 RNA motif is a conserved RNA structure identified by bioinformatics within bacteria in the family bacillaceae.[1] The RNA is presumed to operate as a non-coding RNA, and is sometimes adjacent to operons containing ribosomal RNAs. The most characteristic feature is two terminal loops that have the nucleotide consensus RUCCU, where R is either A or G. The motif might be related to the Desulfotalea-1 RNA motif,[1] as the motifs share some similarity in conserved features, and the Desulfotalea-1 RNA motif is also sometimes adjacent to ribosomal RNA operons.
References
- ↑ 1.0 1.1 "Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea and their metagenomes". Genome Biol 11 (3): R31. March 2010. doi:10.1186/gb-2010-11-3-r31. PMID 20230605.
External links
 |

