Biology:Bacteroides thetaiotaomicron sRNA
| GibS | |
|---|---|
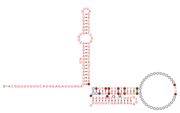
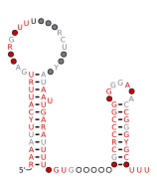
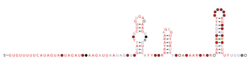 Consensus secondary structure and sequence conservation of GibS sRNA | |
| Identifiers | |
| Symbol | GibS |
| Rfam | RF04182 |
| Other data | |
| RNA type | Gene; sRNA |
| SO | 0000655 |
| PDB structures | PDBe |
The Bacteroides thetaiotaomicron genome contains hundreds of small RNAs (sRNAs),[1] discovered through RNA sequencing. These include canonical housekeeping RNA species such as the 6S RNA (SsrS), tmRNA (SsrA), M1 RNA (RnpB) and 4.5S RNA (Ffs) as well as several hundred cis and trans encoded small RNAs.[1][2] More than 20 candidates have been validated with northern blots and the structures of several members have been characterized through in silico analyses and chemical probing experiments.[1][2]
Two B. thetaiotaomicron sRNAs that have been functionally characterized are RteR and GibS. RteR is a 78 nucleotide (nt) long sRNA that is conserved in closely related species and likely serves as a repressor of a transposon operon.[3][4] Analyses based on secondary structure conservation, taking into consideration nucleotide covariation and in-vitro chemical probing have revealed a structure that consists of a 5’ hairpin and a Rho-independent terminator that are separated by an 8 nt sequence.[3][1] GibS is a 145 nt long sRNA that is also conserved in several closely related species within phylum Bacteroidota and has been hypothesized to play a role in carbon metabolism.[1] Structural analyses have revealed this sRNA to possess an extended 5’ single stranded region (38 nt) followed by two meta-stable hairpins and a Rho-independent terminator at the 3’ end. It is maximally expressed when B. thetaiotaomicron is grown in N-acetyl-D-glucosamine as the sole carbon source and has been shown to both induce and repress target mRNAs involved in metabolic regulation.[1]
The B. thetaiotaomicron genome also contains a large subset of antisense sRNAs that bear resemblance to the B. fragilis DonS RNA.[5] This family of 78 to 128 nt long sRNAs are encoded antisense to several of their target genes, that are members of PULs (Polysaccharide Utilization Loci).[5]
See also
- Bacillus subtilis sRNAs
- Bacterial small RNA
- Brucella sRNA
- Caenorhabditis elegans sRNA
- Escherichia coli sRNA
- Pseudomonas sRNA
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Ryan, Daniel; Jenniches, Laura; Reichardt, Sarah; Barquist, Lars; Westermann, Alexander J. (2020-07-16). "A high-resolution transcriptome map identifies small RNA regulation of metabolism in the gut microbe Bacteroides thetaiotaomicron". Nature Communications 11 (1): 3557. doi:10.1038/s41467-020-17348-5. ISSN 2041-1723. PMID 32678091. Bibcode: 2020NatCo..11.3557R.
- ↑ 2.0 2.1 Prezza, Gianluca; Ryan, Daniel; Mädler, Gohar; Reichardt, Sarah; Barquist, Lars; Westermann, Alexander J. (2021-08-11). "Comparative genomics provides structural and functional insights into Bacteroides RNA biology". Molecular Microbiology 117 (1): 67–85. doi:10.1111/mmi.14793. ISSN 1365-2958. PMID 34379855.
- ↑ 3.0 3.1 Jeters, Robert T.; Wang, Gui-Rong; Moon, Kyung; Shoemaker, Nadja B.; Salyers, Abigail A. (October 2009). "Tetracycline-associated transcriptional regulation of transfer genes of the Bacteroides conjugative transposon CTnDOT". Journal of Bacteriology 191 (20): 6374–6382. doi:10.1128/JB.00739-09. ISSN 1098-5530. PMID 19700528.
- ↑ Waters, Jillian L.; Salyers, Abigail A. (October 2012). "The small RNA RteR inhibits transfer of the Bacteroides conjugative transposon CTnDOT". Journal of Bacteriology 194 (19): 5228–5236. doi:10.1128/JB.00941-12. ISSN 1098-5530. PMID 22821972.
- ↑ 5.0 5.1 Cao, Yanlu; Förstner, Konrad U.; Vogel, Jörg; Smith, C. Jeffrey (2016-09-15). "cis-Encoded Small RNAs, a Conserved Mechanism for Repression of Polysaccharide Utilization in Bacteroides". Journal of Bacteriology 198 (18): 2410–2418. doi:10.1128/JB.00381-16. ISSN 1098-5530. PMID 27353652.
 |