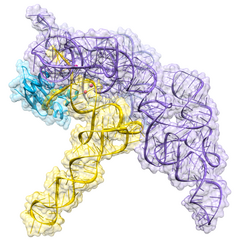
Biology:Ribonuclease P
| Bacterial RNase P class A | |
|---|---|
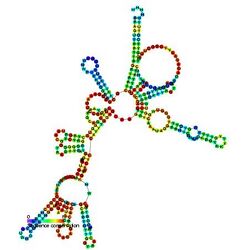 Predicted secondary structure and sequence conservation of RNaseP_bact_a | |
| Identifiers | |
| Symbol | RNaseP_bact_a |
| Rfam | RF00010 |
| Other data | |
| RNA type | Gene; ribozyme |
| Domain(s) | Bacteria |
| GO | 0008033 0004526 0030680 |
| SO | 0000386 |
| PDB structures | PDBe |
| Bacterial RNase P class B | |
|---|---|
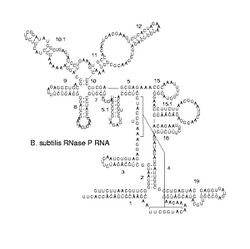 Predicted secondary structure and sequence conservation of RNaseP_bact_b | |
| Identifiers | |
| Symbol | RNaseP_bact_b |
| Rfam | RF00011 |
| Other data | |
| RNA type | Gene; ribozyme |
| Domain(s) | Bacteria |
| GO | 0008033 0004526 0030680 |
| SO | 0000386 |
| PDB structures | PDBe |
| Archaeal RNase P | |
|---|---|
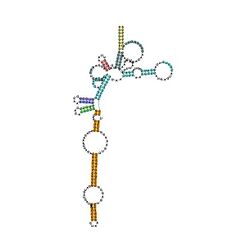 Predicted secondary structure and sequence conservation of Archaeal RNase P | |
| Identifiers | |
| Symbol | RNaseP_arch |
| Rfam | RF00373 |
| Other data | |
| RNA type | Gene; ribozyme |
| Domain(s) | Archaea |
| GO | 0008033 0004526 0030680 |
| SO | 0000386 |
| PDB structures | PDBe |
| Archaeal RNase P class T | |
|---|---|
| Identifiers | |
| Symbol | RNaseP-T |
| Rfam | RF02357 |
| Other data | |
| RNA type | Gene; ribozyme |
| Domain(s) | Archaea |
| GO | 0008033 0004526 0030680 |
| SO | 0000386 |
| PDB structures | PDBe |
Ribonuclease P (EC 3.1.26.5, RNase P) is a type of ribonuclease which cleaves RNA. RNase P is unique from other RNases in that it is a ribozyme – a ribonucleic acid that acts as a catalyst in the same way that a protein-based enzyme would. Its function is to cleave off an extra, or precursor, sequence of RNA on tRNA molecules.[1] Further, RNase P is one of two known multiple turnover ribozymes in nature (the other being the ribosome), the discovery of which earned Sidney Altman and Thomas Cech the Nobel Prize in Chemistry in 1989: in the 1970s, Altman discovered the existence of precursor tRNA with flanking sequences and was the first to characterize RNase P and its activity in processing of the 5' leader sequence of precursor tRNA. Recent findings also reveal that RNase P has a new function.[2] It has been shown that human nuclear RNase P is required for the normal and efficient transcription of various small noncoding RNAs, such as tRNA, 5S rRNA, SRP RNA and U6 snRNA genes,[3] which are transcribed by RNA polymerase III, one of three major nuclear RNA polymerases in human cells.
In Bacteria
Bacterial RNase P has two components: an RNA chain, called M1 RNA, and a polypeptide chain, or protein, called C5 protein.[4][5] In vivo, both components are necessary for the ribozyme to function properly, but in vitro, the M1 RNA can act alone as a catalyst.[1] The primary role of the C5 protein is to enhance the substrate binding affinity and the catalytic rate of the M1 RNA enzyme probably by increasing the metal ion affinity in the active site. The crystal structure of a bacterial RNase P holoenzyme with tRNA has been recently resolved, showing how the large, coaxially stacked helical domains of the RNase P RNA engage in shape selective recognition of the pre-tRNA target. This crystal structure confirms earlier models of substrate recognition and catalysis, identifies the location of the active site, and shows how the protein component increases RNase P functionality.[6][7]
Bacterial RNase P class A and B
Ribonuclease P (RNase P) is a ubiquitous endoribonuclease, found in archaea, bacteria and eukarya as well as chloroplasts and mitochondria. Its best characterised activity is the generation of mature 5'-ends of tRNAs by cleaving the 5'-leader elements of precursor-tRNAs. Cellular RNase Ps are ribonucleoproteins (RNP). RNA from bacterial RNase Ps retains its catalytic activity in the absence of the protein subunit, i.e. it is a ribozyme. Isolated eukaryotic and archaeal RNase P RNA has not been shown to retain its catalytic function, but is still essential for the catalytic activity of the holoenzyme. Although the archaeal and eukaryotic holoenzymes have a much greater protein content than the eubacterial ones, the RNA cores from all the three lineages are homologous—helices corresponding to P1, P2, P3, P4, and P10/11 are common to all cellular RNase P RNAs. Yet, there is considerable sequence variation, particularly among the eukaryotic RNAs.
In Archaea
In archaea, RNase P ribonucleoproteins consist of 4–5 protein subunits that are associated with RNA. As revealed by in vitro reconstitution experiments these protein subunits are individually dispensable for tRNA processing that is essentially mediated by the RNA component.[8][9][10] The structures of protein subunits of archaeal RNase P have been resolved by x-ray crystallography and NMR, thus revealing new protein domains and folding fundamental for function.
Using comparative genomics and improved computational methods, a radically minimized form of the RNase P RNA, dubbed "Type T", has been found in all complete genomes in the crenarchaeal phylogenetic family Thermoproteaceae, including species in the genera Pyrobaculum, Caldivirga and Vulcanisaeta.[11] All retain a conventional catalytic domain, but lack a recognizable specificity domain. 5′ tRNA processing activity of the RNA alone was experimentally confirmed. The Pyrobaculum and Caldivirga RNase P RNAs are the smallest naturally occurring form yet discovered to function as trans-acting ribozymes.[11] Loss of the specificity domain in these RNAs suggests potential altered substrate specificity.
It has recently been argued that the archaebacteriium Nanoarchaeum equitans does not possess RNase P. Computational and experimental studies failed to find evidence for its existence. In this organism the tRNA promoter is close to the tRNA gene and it is thought that transcription starts at the first base of the tRNA thus removing the requirement for RNase P.[12]
In eukaryotes
In eukaryotes, such as humans and yeast,[13] most RNase P consists of an RNA chain that is structurally similar to that found in bacteria [14] as well as nine to ten associated proteins (as opposed to the single bacterial RNase P protein, C5).[2][15] Five of these protein subunits exhibit homology to archaeal counterparts. These protein subunits of RNase P are shared with RNase MRP,[15][16][17] a catalytic ribonucleoprotein involved in processing of ribosomal RNA in the nucleolus.[18] RNase P from eukaryotes was only recently demonstrated to be a ribozyme.[19] Accordingly, the numerous protein subunits of eucaryal RNase P have a minor contribution to tRNA processing per se,[20] while they seem to be essential for the function of RNase P and RNase MRP in other biological settings, such as gene transcription and the cell cycle.[3][21] Despite the bacterial origins of mitochondria and chloroplasts, plastids from higher animals and plants do not appear to contain an RNA-based RNase P. It has been shown that human mitochondrial RNase P is a protein and does not contain RNA.[22] Spinach chloroplast RNase P has also been shown to function without an RNA subunit.[23]
| Subunit | Function/interaction (in tRNA processing) |
|---|---|
| RPP14 | RNA binding |
| RPP20 | ATPase, helicase/Hsp27, SMN, Rpp25 |
| RPP21 | RNA binding, activityg/Rpp29 |
| RPP25 | RNA binding/Rpp20 |
| RPP29 | tRNA binding, activity/Rpp21 |
| RPP30 | RNA binding, activity/Pop5 |
| RPP38 | RNA binding, activity |
| RPP40 | |
| hPop1 | |
| hPop5 | RNA binding, activity/Rpp30 |
| H1 RNA | Activity/Rpp21, Rpp29, Rpp30, Rpp38 |
Therapies using RNase P
RNase P is now being studied as a potential therapy for diseases such as herpes simplex virus,[24] cytomegalovirus,[24][25] influenza and other respiratory infections,[26] HIV-1[27] and cancer caused by fusion gene BCR-ABL.[24][28] External guide sequences (EGSs) are formed with complementarity to viral or oncogenic mRNA and structures that mimic the T loop and acceptor stem of tRNA.[26] These structures allow RNase P to recognize the EGS and cleave the target mRNA. EGS therapies have shown to be effective in culture and in live mice.[29]
References
- ↑ 1.0 1.1 "The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme". Cell 35 (3 Pt 2): 849–57. December 1983. doi:10.1016/0092-8674(83)90117-4. PMID 6197186.
- ↑ 2.0 2.1 2.2 "Human RNase P: a tRNA-processing enzyme and transcription factor". Nucleic Acids Research 35 (11): 3519–24. 2007. doi:10.1093/nar/gkm071. PMID 17483522.
- ↑ 3.0 3.1 "A role for the catalytic ribonucleoprotein RNase P in RNA polymerase III transcription". Genes & Development 20 (12): 1621–35. June 2006. doi:10.1101/gad.386706. PMID 16778078.
- ↑ "RNase P: interface of the RNA and protein worlds". Trends in Biochemical Sciences 31 (6): 333–41. June 2006. doi:10.1016/j.tibs.2006.04.007. PMID 16679018.
- ↑ "Molecular modeling of the three-dimensional structure of the bacterial RNase P holoenzyme". Journal of Molecular Biology 325 (4): 661–75. January 2003. doi:10.1016/S0022-2836(02)01267-6. PMID 12507471. http://www-ibmc.u-strasbg.fr/upr9002/westhof/PDF/2003_HTsai_JMB.pdf.
- ↑ "Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA". Nature 468 (7325): 784–9. December 2010. doi:10.1038/nature09516. PMID 21076397. Bibcode: 2010Natur.468..784R.
- ↑ "RNase P: at last, the key finds its lock". RNA 17 (9): 1615–8. September 2011. doi:10.1261/rna.2841511. PMID 21803972.
- ↑ "Archaeal RNase P has multiple protein subunits homologous to eukaryotic nuclear RNase P proteins". RNA 8 (3): 296–306. March 2002. doi:10.1017/S1355838202028492. PMID 12003490.
- ↑ "A fifth protein subunit Ph1496p elevates the optimum temperature for the ribonuclease P activity from Pyrococcus horikoshii OT3". Biochemical and Biophysical Research Communications 343 (3): 956–64. May 2006. doi:10.1016/j.bbrc.2006.02.192. PMID 16574071.
- ↑ "Functional reconstitution and characterization of Pyrococcus furiosus RNase P". Proceedings of the National Academy of Sciences of the United States of America 103 (44): 16147–52. October 2006. doi:10.1073/pnas.0608000103. PMID 17053064. Bibcode: 2006PNAS..10316147T.
- ↑ 11.0 11.1 "Discovery of a minimal form of RNase P in Pyrobaculum". Proceedings of the National Academy of Sciences of the United States of America 107 (52): 22493–8. December 2010. doi:10.1073/pnas.1013969107. PMID 21135215. Bibcode: 2010PNAS..10722493L.
- ↑ "Life without RNase P". Nature 453 (7191): 120–3. May 2008. doi:10.1038/nature06833. PMID 18451863. Bibcode: 2008Natur.453..120R.
- ↑ Randall Munroe rephrased this as “You know, eukaryotes—like sourdough starter or Conan O’Brien.” (Munroe, Randall (30 September 2022). "4:25 PM". https://twitter.com/xkcd/status/1575944901090455552.)
- ↑ "Structure and function of eukaryotic Ribonuclease P RNA". Molecular Cell 24 (3): 445–56. November 2006. doi:10.1016/j.molcel.2006.09.011. PMID 17081993.
- ↑ 15.0 15.1 "Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP". Genes & Development 12 (11): 1678–90. June 1998. doi:10.1101/gad.12.11.1678. PMID 9620854.
- ↑ "Characterization and purification of Saccharomyces cerevisiae RNase MRP reveals a new unique protein component". The Journal of Biological Chemistry 280 (12): 11352–60. March 2005. doi:10.1074/jbc.M409568200. PMID 15637077.
- ↑ "Differential association of protein subunits with the human RNase MRP and RNase P complexes". RNA 12 (7): 1373–82. July 2006. doi:10.1261/rna.2293906. PMID 16723659.
- ↑ "A big development for a small RNA". Nature 410 (6824): 29–31. March 2001. doi:10.1038/35065191. PMID 11242026.
- ↑ "Eukaryotic RNase P RNA mediates cleavage in the absence of protein". Proceedings of the National Academy of Sciences of the United States of America 104 (7): 2062–7. February 2007. doi:10.1073/pnas.0607326104. PMID 17284611.
- ↑ "An important piece of the RNase P jigsaw solved". Trends in Biochemical Sciences 32 (6): 247–50. June 2007. doi:10.1016/j.tibs.2007.04.005. PMID 17485211.
- ↑ "RNase MRP cleaves the CLB2 mRNA to promote cell cycle progression: novel method of mRNA degradation". Molecular and Cellular Biology 24 (3): 945–53. February 2004. doi:10.1128/MCB.24.3.945-953.2004. PMID 14729943.
- ↑ "RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme". Cell 135 (3): 462–74. October 2008. doi:10.1016/j.cell.2008.09.013. PMID 18984158.
- ↑ "Chloroplast ribonuclease P does not utilize the ribozyme-type pre-tRNA cleavage mechanism". RNA 6 (4): 545–53. April 2000. doi:10.1017/S1355838200991465. PMID 10786845.
- ↑ 24.0 24.1 24.2 "Developing RNase P ribozymes for gene-targeting and antiviral therapy". Cellular Microbiology 6 (6): 499–508. June 2004. doi:10.1111/j.1462-5822.2004.00398.x. PMID 15104592.
- ↑ "RNase P ribozymes for the studies and treatment of human cytomegalovirus infections". Journal of Clinical Virology 25 Suppl 2: S63-74. August 2002. doi:10.1016/s1386-6532(02)00097-5. PMID 12361758.
- ↑ 26.0 26.1 "Gene silencing in the therapy of influenza and other respiratory diseases: Targeting to RNase P by use of External Guide Sequences (EGS)". Biologics: Targets and Therapy 1 (4): 425–32. December 2007. PMID 19707312.
- ↑ "Effective inhibition of human immunodeficiency virus 1 replication by engineered RNase P ribozyme". PLOS ONE 7 (12): e51855. December 26, 2012. doi:10.1371/journal.pone.0051855. PMID 23300569. Bibcode: 2012PLoSO...751855Z.
- ↑ "In vivo inhibition by a site-specific catalytic RNA subunit of RNase P designed against the BCR-ABL oncogenic products: a novel approach for cancer treatment". Blood 95 (3): 731–7. February 2000. doi:10.1182/blood.V95.3.731.003k28_731_737. PMID 10648380.
- ↑ "A peptide-morpholino oligomer conjugate targeting Staphylococcus aureus gyrA mRNA improves healing in an infected mouse cutaneous wound model". International Journal of Pharmaceutics 453 (2): 651–5. September 2013. doi:10.1016/j.ijpharm.2013.05.041. PMID 23727592.
Further reading
- "Ribonuclease P: unity and diversity in a tRNA processing ribozyme". Annual Review of Biochemistry 67: 153–80. 1998. doi:10.1146/annurev.biochem.67.1.153. PMID 9759486.
- "The Ribonuclease P Database". Nucleic Acids Research 27 (1): 314. January 1999. doi:10.1093/nar/27.1.314. PMID 9847214.
External links
- Nobel Lecture of Sidney Altman, Nobel prize in Chemistry 1989
- RNase P Database at ncsu.edu
- Page for Nuclear RNase P at Rfam
- Page for Archaeal RNase P at Rfam
- Page for Bacterial RNase P class A at Rfam
- Page for Bacterial RNase P class B at Rfam
- RNase+P at the US National Library of Medicine Medical Subject Headings (MeSH)
- EC 3.1.26.5
 |