Biology:C9orf85
 Generic protein structure example |
Chromosome 9 open reading frame 85, commonly known as C9orf85, is a protein in Homo sapiens encoded by the C9orf85 gene. The gene is located at 9q21.13.[1] When spliced, four different isoforms are formed. C9orf85 has a predicted molecular weight of 20.17 kdal.[2] Isoelectric point was found to be 9.54.[2] The function of the gene has not yet been confirmed, however it has been found to show high levels of expression in cells of high differentiation.[3]
Background
Protein Sequence
The sequence for C9orf85 isoform 1 in Homo sapiens, derived from NCBI:[1]
MSSQKGNVARSRPQKHQNTFSFKNDKFDKSVQTKKINAKLHDGVCQRCKEVLEWRVKYSKYKPLSKPKKCVKCLQKTVKDSYHIMCRPCACELEVCAKCGKKEDIVIPWSLPLLPRLECSGRILAHHNLRLPCSSDSPAS ASRVAGTTGAHHHAQLIFVFLVEMGFHYVGQAGLELLTS
Aliases
Isoforms
| Isoform # | mRNA Length (bp) | Amino Acid Length (aa) |
|---|---|---|
| 1[7] | 3821 | 179 |
| 2[8] | 1185 | 157 |
| 3[9] | 1316 | 138 |
| 4[10] | 3707 | 69 |
Isoform 1 is the major form of the gene used. This isoform contains 4 exons. It's accession number is NM_001365053.2.[1]
Homology
Orthologs
The C9orf85 gene was found in all species type including vertebrate to bacteria. However no type of protist was found as an ortholog for the human gene except for plasmodium.
| Genus species | Common Name | Taxonomic Group | Date of Divergence (MYA) | Accession Number | Length (aa) | Identity | Similarity |
|---|---|---|---|---|---|---|---|
| Homo sapiens | Human | Chordata | 0 | NP_001351982 | 179 | 100% | 100% |
| Meriones unguiculatus | Mongolian gerbil | Rodentia | 90 | XP_021514638 | 154 | 74% | 84% |
| Gallus gallus | Chicken | Chordata | 312 | XP_001233821 | 166 | 78.70% | 60% |
| Terrapene carolina triunguis | Three-toed box turtle | Chordata | 312 | XP_024066792 | 171 | 85.45% | 61% |
| Chelonia mydas | Green sea turtle | Chordata | 312 | XP_007065676 | 178 | 84.31% | 61% |
| Calidris pugnax | Ruff bird | Chordata | 312 | XP_014813985 | 166 | 77.78% | 60% |
| Microcaecilia unicolor | Tiny cayenne caecilian | Chordata | 351.8 | XP_030049723 | 178 | 76.15% | 60% |
| Xenopus tropicalis | Western clawed frog | Chordata | 351.8 | KAE8633085 | 133 | 78.18% | 61% |
| Electrophorus electricus | Electric eel | Chordata | 435 | XP_026886158 | 156 | 55.13% | 87% |
| Oncorhynchus mykiss | Rainbow trout | Chordata | 435 | XP_021461156 | 177 | 60.77% | 72% |
| Acanthaster planci | Crown-of-thorns starfish | Echinodermata | 684 | XP_022096254 | 197 | 56.76% | 62% |
| Photinus pyralis | Big dipper firefly | Arthropoda | 797 | XP_031346726 | 183 | 58.04% | 62% |
| Pomacea canaliculata | Golden apple snail | Mollusca | 797 | XP_025077101 | 208 | 47.33% | 73% |
| Drosophila melanogaster | Fruit Fly | Arthropoda | 797 | NP_573209 | 234 | 49.58% | 65% |
| Acropora millepora | Coral | Cnidaria | 824 | XP_029187517 | 190 | 57.14% | 62% |
| Salpingoeca rosetta | Choanoflagellate | Choanoflagellate | 1023 | XP_004995700 | 286 | 41.67% | 60% |
| Apophysomyces ossiformis | Fungi | Mucoromycota | 1105 | KAF7725139 | 181 | 40% | 72% |
| Ricinus communis | Caster oil plant | Spermatophyta | 1496 | XP_002530997 | 227 | 34.21% | 78% |
| Plasmodium ovale wallikeri | Malarian protist | Apicomplexa | 1768 | SBT56954 | 680 | 68.75% | 17% |
| Bacillus cereus | Bacteria | Firmicutes | 4290 | KXI72539 | 83 | 73.61% | 39% |
Paralogs
| Paralog | Accession Number | Length (aa) | Identity | Similarity | Location |
|---|---|---|---|---|---|
| CCDC198 | XP_005267863 | 290 | 44.38% | 89% | Chromosome 14 |
| Retbidin | EAW84316 | 224 | 60% | 51% | Chromosome 19 |
| hCG2038446 | EAX11460 | 135 | 68.54% | 49% | Chromosome 2 |
| hCG1820974 | EAW94215 | 143 | 72.58% | 41% | Chromosome 17 |
| O-phosphoseryl-tRNA(Sec) selenium transferase isoform X1 | XP_016863766 | 586 | 70.67% | 41% | Chromosome 4 |
Rate of Molecular Evolution
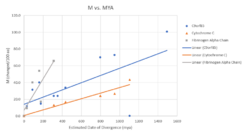
A rate of divergence can be calculated using the molecular clock hypothesis. As observed by the graph, C9orf85 lies between Cytochrome C and Fibrinogen Alpha with a slope leaning more towards Cytochrome C. Therefore, C9orf85 is possibly evolving at a slower rate than most proteins.
Conservation
Multiple Sequence Alignment
A multiple sequence alignment (MSA)[12] was done between the top 15 closely related orthologs to the Homo sapiens C9orf85. 20 amino acids were discovered to be conserved among all 15 sequences at the beginning of the protein sequence; within the first three exons.
In a MSA between distantly related homologs, 5 amino acids were observed to be conserved between exons two and three.
Yet, when running a multiple sequence alignment between Homo sapiens and the extremely distant Bacillus cereus, 53 amino acids are observed to be conserved primarily in the fourth exon.
Cysteine
The amino acid cysteine appears the most throughout the protein sequence as a conserved amino acids; 8 out of 20 instances. Cysteine 48, 70, 89, 96, and Tryptophan 54 are amino acids conserved in all species type – including vertebrate, invertebrate, fungi, plants, and protists – besides bacteria.
Using the Statistical Analysis of Protein Sequences tool,[2] SAPS, 5 spacings of cysteine were found. Four with the pattern of C-X-X-C—at amino acids 45, 70, 86, and 96—and the fifth spacing at amino acid 89 (CAC). The C-X-X-C pattern is known to be present in metal-binding proteins and oxidoreductases.[13] Additionally, three of the five cysteine spacings were also the top conserved amino acids throughout the most closely related orthologs; C70, C89, and C96.
Localization
Gene Localization in Humans
C9orf85 has been found to be expressed highly in epithelial cells.[14] of the pancreas.[15] Additionally, high levels of expression have been established in the urinary bladder and thymus of the adult human, while expression levels were significant in the intestine of a 20-week-old fetus.[1]
Subcellular Localization
k-NN results predict C9orf85 to be 78.3% nuclear, 8.7% mitochondrial, 8.7% cytoplasmic, and 4.3% vacuolar.[16]
Promoter
C9orf85 has 3 predicted promoters for the gene.[17] The choice promoter was GXG_18858 on the plus strand. Chosen for its large quantity of CAGE tags and its position being furthest upstream. Its start position is 71909780 and its end position is 71911841. It includes 2062 base pairs and has 13 transcripts. The last 500 base pairs of the double stranded promoter is featured below:
5' GCAGGAGGCGGGGATTGCGGAAAAGAAGAACCAATAGGAACAAAGGTTCC 3' 3' CGTCCTCCGCCCCTAACGCCTTTTCTTCTTGGTTATCCTTGTTTCCAAGG 5'
5' CCGCCCCTTTGATTTGATGGACTACACATTCGGGCCAATGGGGGAATTCT 3' 3' GGCGGGGAAACTAAACTACCTGATGTGTAAGCCCGGTTACCCCCTTAAGA 5'
5' CATTTCGAAGAAAGTGGGACTTGTTCTCCGGGTTTGAGAAAGAGGCTGCG 3' 3' GTAAAGCTTCTTTCACCCTGAACAAGAGGCCCAAACTCTTTCTCCGACGC 5'
5' CGGAGCCGGAGGGGTCGAGGCTGCGCCGCGTGGAGTGGCTTGGCTTAACA 3' 3' GCCTCGGCCTCCCCAGCTCCGACGCGGCGCACCTCACCGAACCGAATTGT 5'
5' GCAGGGAGGGCAGAGCGATGCTCTTTGACCTCCCAGAAGAGTCACGTGGG 3' 3' CGTCCCTCCCGTCTCGCTACGAGAAACTGGAGGGTCTTCTCAGTGCACCC 5'
5' CTGACCCAGAGCCGGGGCGGAAAGGCTGCGTTTGTTTCTTCCGGGTCATT 3' 3' GACTGGGTCTCGGCCCCGCCTTTCCGACGCAAACAAAGAAGGCCCAGTAA 5'
5' GACAGAAGCGTCAATTCCTGGGAGTAGTTCGTTGGTTTTCTTTCCCCTCA 3' 3' CTGTCTTCGCAGTTAAGGACCCTCATCAAGCAACCAAAAGAAAGGGGAGT 5'
5' TCCTTTTGCCTGCTCCCGGCGAGGGGTGGCTTTGATTTCGGCGATGAGCT 3' 3' AGGAAAACGGACGAGGGCCGCTCCCCACCGAAACTAAAGCCGCTACTCGA 5'
5' CCCAGAAAGGCAACGTGGCTCGTTCCAGACCTCAGAAGCACCAGAATACG 3' 3' GGGTCTTTCCGTTGCACCGAGCAAGGTCTGGAGTCTTCGTGGTCTTATGC 5'
5' TTTAGCTTCAAAAATGACAAGTTCGATAAAAGTGTGCAGACCAAGGTAGG 3' 3' AAATCGAAGTTTTTACTGTTCAAGCTATTTTCACACGTCTGGTTCCATCC 5'
| Transcription Factor | Detailed Matrix Information | Matrix Score |
|---|---|---|
| CLOX | Transcriptional repressor CDP | 0.962 |
| KLFS | Gut-enriched Krueppel-like factor | 1.000 |
| CAAT | Nuclear factor Y (Y-box binding factor) | 0.940 |
| HIFF | Aryl hydrocarbon receptor nuclear translocator-like, homodimer | 1.000 |
| MZF1 | Myeloid zinc finger protein | 0.992 |
| STAT | STAT5: signal transducer and activator of transcription 5 | 0.944 |
| ETSF | ETS-like gene 1 (ELK-1) | 0.958 |
| CREB | Tax/CREB complex | 0.834 |
| P53F | Tumor suppressor p53 (3' half site) | 0.921 |
| TCFF | TCF11/LCP-F1/Nrf1 homodimers | 1.000 |
| FKHD | Fork head homologous X binds DNA with a dual sequence specificity (FHXA and FHXB) | 0.870 |
| MIRF | Zinc finger protein 768 | 0.819 |
| BCL6 | B-cell CLL/lymphoma 6, member B (BCL6B) | 0.878 |
| AP2F | Transcription factor AP-2, alpha | 0.931 |
| EBOX | MYC associated factor X | 0.926 |
| GCMF | Glial cells missing homolog 1, chorion-specific transcription factor GCMa | 0.942 |
Regulation
Transmembrane Domain
Though there is a presence of hydrophobic regions in the protein sequence,[2][19][20] there have been no confirmed transmembrane domains present[21]
Phosphorylation
A protein kinase C phosphorylation site is predicted at amino acid 3-5.[22] There is also a possible CK2 phosphorylation site at amino acid 77-80[22]
SUMOylating
There is one predicted SUMO site at position 23.[23] The result is significant with a p-value of 0.041.
Function
Through the level of expression in various tissue samples, the C9orf85 protein is a regulated gene rather than a constitutive gene.[1]
Additionally, urinary bladder epithelial cells function by altering the immune system of an infection.[24] The thymus is a primary lymphoid organ of the immune system, composed of T cells and epithelial cells. Research has found that the thymus has an increasing role in the development of intestinal immunity[25] Both are an element of the immune system, designed to ensure proper function of the immune system.
References
- ↑ 1.0 1.1 1.2 1.3 1.4 "C9orf85 chromosome 9 open reading frame 85 [Homo sapiens (human) – Gene – NCBI"]. https://www.ncbi.nlm.nih.gov/gene/138241.
- ↑ 2.0 2.1 2.2 2.3 EMBL-EBI. (2020). SAPS Results. Ebi.Ac.Uk. https://www.ebi.ac.uk/Tools/services/web/toolresult.ebi?jobId=saps-I20201219-191317-0344-54841082-p1m
- ↑ "Identification of microRNAs expressed highly in pancreatic islet-like cell clusters differentiated from human embryonic stem cells". Cell Biology International 35 (1): 29–37. January 2011. doi:10.1042/CBI20090081. PMID 20735361.
- ↑ C9orf85 - Uncharacterized protein C9orf85 - Homo sapiens (Human) - C9orf85 gene & protein. (2020). Uniprot.Org. https://www.uniprot.org/uniprot/Q96MD7
- ↑ (2020). Genenames.Org. https://www.genenames.org/data/gene-symbol-report/#!/hgnc_id/28784
- ↑ 6.0 6.1 6.2 "AceView a comprehensive annotation of human and worm genes with mRNAs or ESTsAceView.". https://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/.
- ↑ "uncharacterized protein C9orf85 isoform 1 [Homo sapiens – Protein – NCBI"]. https://www.ncbi.nlm.nih.gov/protein/NP_001351982.1.
- ↑ "uncharacterized protein C9orf85 isoform 2 [Homo sapiens – Protein – NCBI"]. https://www.ncbi.nlm.nih.gov/protein/NP_872311.2.
- ↑ "uncharacterized protein C9orf85 isoform 3 [Homo sapiens – Protein – NCBI"]. https://www.ncbi.nlm.nih.gov/protein/NP_001351984.1.
- ↑ "uncharacterized protein C9orf85 isoform 4 [Homo sapiens – Protein – NCBI"]. https://www.ncbi.nlm.nih.gov/protein/NP_001351986.1.
- ↑ 11.0 11.1 "Protein BLAST: search protein databases using a protein query". https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome.
- ↑ "Clustal Omega < Multiple Sequence Alignment < EMBL-EBI". https://www.ebi.ac.uk/Tools/msa/clustalo/.
- ↑ "Relationship between the occurrence of cysteine in proteins and the complexity of organisms". Molecular Biology and Evolution 17 (8): 1232–9. August 2000. doi:10.1093/oxfordjournals.molbev.a026406. PMID 10908643.
- ↑ GENEVESTIGATOR Team at Nebion AG. (2020). Genevisible. Genevisible.com; genevisible. https://genevisible.com/tissues/HS/UniProt/Q96MD7
- ↑ "Gene: C9orf85 – ENSG00000155621" (in en). https://bgee.org/?page=gene&gene_id=ENSG00000155621.
- ↑ PSORT II Prediction. (2020). Psort.Hgc.Jp. https://psort.hgc.jp/form2.html
- ↑ Genomatix: Gene2Promoter Result. (2020). Genomatix.De. https://www.genomatix.de/cgi-bin/c2p/c2p.pl?s=c5402bf929e4d6000dfc7ce8c56fa1e6;TASK=c2p;SHOW=TempSeq_kd0ZKohP.html
- ↑ Genomatix: MatInspector Result. (2019). Genomatix.De. https://www.genomatix.de/cgi-bin/eldorado/eldorado.pl?s=c5402bf929e4d6000dfc7ce8c56fa1e6;PROM_ID=GXP_18858;GROUP=vertebrates;GROUP=others;ELDORADO_VERSION=E35R1911
- ↑ "ProtScale". https://web.expasy.org/cgi-bin/protscale/protscale.pl?1.
- ↑ TMPred results. (2020). Vital-It.Ch. https://embnet.vital-it.ch/cgi-bin/TMPRED_form_parser
- ↑ "SOSUI/submit a protein sequence". http://harrier.nagahama-i-bio.ac.jp/sosui/sosui_submit.html.
- ↑ 22.0 22.1 "Motif Scan" (in en). https://myhits.sib.swiss/cgi-bin/motif_scan.
- ↑ "GPS-SUMO: Prediction of SUMOylation Sites & SUMO-interaction Motifs". http://sumosp.biocuckoo.org/showResult.php.
- ↑ "The nature of immune responses to urinary tract infections". Nature Reviews. Immunology 15 (10): 655–63. October 2015. doi:10.1038/nri3887. PMID 26388331.
- ↑ "A ticket to the gut for thymic T cells". Gut 55 (7): 910–2. July 2006. doi:10.1136/gut.2005.087288. PMID 16766746.
 |