Biology:CCDC47
 Generic protein structure example |
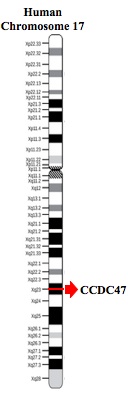
Coiled-coil domain 47 (CCDC47) is a gene located on human chromosome 17, specifically locus 17q23.3 which encodes for the protein CCDC47. The gene has several aliases including GK001 and MSTP041. The protein itself contains coiled-coil domains, the SEEEED superfamily, a domain of unknown function (DUF1682) and a transmembrane domain. The function of the protein is unknown, but it has been proposed that CCDC47 is involved in calcium ion homeostasis and the endoplasmic reticulum overload response.[1]
Gene
The CCDC47 gene itself is located on the minus strand of human chromosome 17 and contains 13 exon splice sites and 14 distinct introns. After removal of exons, the gene is 3445 base pairs in length. No evidence for micro RNA or pseudogenes has been found. The gene does not have various isoforms, only transcript variant 1X exists.
Protein
Structure
The protein encoded by CCDC47 is 483 amino acids in length and contains both a signal peptide and transmembrane domain. It is rich in negatively charged amino acids such as aspartic acid and glutamic acid giving it an acidic isoelectric point of 4.56.[3] The protein is also rich in methionine. In total, it weighs 55.9 kDal which is conserved through various orthologs. CCDC47 also contains the SEEEED superfamily and domain of unknown function 1682 (DUF1682). The SEEEED superfamily is a short, low complexity region which is composed mainly of serine. The family routinely lies on the clathrin adaptor complex 3 beta-1 subunit proteins.[4] The exact function of DUF 1682 is unclear but one member of the family has been described as an adipocyte-specific protein.[5]
There are two predicted disulfide bonds in the structure of CCDC47 at cysteines 209 to 214 and cysteines 215 to 283, respectively.[6] The C-terminal portion of the protein is highly charged and its secondary structure is predicted to be that of an alpha helix region.[7] This region also contains coiled coil domains which are structural motifs in which 2-7 alpha helices are coiled together and are subsequently involved in biological expression. These domains typically follow the pattern HxxHCxC where H is a hydrophobic amino acid, C is a charged amino acid and x is any amino acid.[8] Many amino acid sequences following this pattern are seen in the C-terminal region of CCDC47 where the highest conservation through orthologs is represented.

Regulation and translation
CCDC47 is regulated by the promoter GXP43413.[9] The promoter is 819 base pairs in length and is highly conserved in mammals. Conserved binding sites in mammals which are located on this promoter include Nuclear Respiratory Factor 1 (NFR1), cAMP-responsive element binding protein (CREB), PAR b ZIP family and Sp4 Transcription Factor. NRF1 encodes a protein which homodimerizes and activates expression of key metabolic genes. CREB binds to cAMP response elements thereby increasing or decreasing the transcription of downstream genes[10] while PAR b ZIP family is involved in the regulation of circadian rhythms.[11] In regards to the mRNA, translation begins at base pair 337 and ends at 1728. There is a strong stem loop located in the 5' UTR region from bases 289-318 which likely is involved in regulation of the mRNA due to its close proximity to the start codon.[12]
Cellular distribution
The final protein is thought to be translated from the endoplasmic reticulum into the cytoplasm of the cell. The protein is anchored in the membrane of the ER at the transmembrane domain located from amino acid 137 to 165.[13] The portion of the protein which extends into the cytosol is predicted to be highly phosphorylated as the protein's phosphorylation sites are conserved into the bony fish orthologs.[14] Research has shown that CCDC47 is expressed in the response to an ER overload making this close proximity to the ER important.[15]
Post translational modification
In addition to the high levels of phosphorylation seen in CCDC47, three sulfation sites are predicted and conserved in mammals, reptiles and birds but not in fish, amphibians or invertebrates.[16] Five potential sumoylation sites are also seen and conserved back to the bony fish.[17] There is no glycosylation of the protein as it is not predicted to extend into the extracellular portion of the cell.
Expression
Microarray tissue expression patterns from GEO were analyzed and showed that CCDC47 appears to be an ubiquitously expressed at moderate levels in many different human tissues.[18] Although the protein is ubiquitously expressed, the highest levels of expression are seen in neuronal tissues such as the superior cervical ganglion, brain amygdala and ciliary ganglion. Elevated expression is also seen in the thyroid and CD34+ cells.
Homology
CCDC47 has no known paralogs through text based queries, BLAST and BLAT. The gene has many orthologs extending back to invertebrates such as C. elegans and is highly conserved in mammals with a percent identity greater than 95%. CCDC47 has been sequenced in a wide taxonomy of organisms including mammals, birds, reptiles, amphibians, bony fish and invertebrates. Percent identity of human CCDC47 to a specific ortholog declines with increasing years of divergence, as expected. Homologous genes of CCDC47 are also present in mosquitos, mushrooms, arabidopsis and Asian rice. These homologs contain the same DUF1682 which is found in CCDC47.
| Genus
Species |
Common Organism Name | Divergence from
Humans (MYA)[19] |
NCBI Protein
Accession Number |
Sequence Identity
to Humans[20] |
Sequence Length
(AA) |
|---|---|---|---|---|---|
| Mus musculus | Mouse | 92.3 | NP_080285.2 | 97.90% | 483 |
| Myotis davidii | Mouse-eared Bat | 94.2 | XP_006776781.1 | 97.50% | 483 |
| Elephantulus edwardii | Elephant Shrew | 98.7 | XP_006886355.1 | 95.00% | 483 |
| Alligator mississippiensis | American Alligator | 296 | XP_006271625.1 | 91.00% | 482 |
| Falco cherrug | Saker Falcon | 296 | XP_005439470.1 | 90.10% | 482 |
| Ophiophagus hannah | King Cobra | 296 | ETE73955 | 78.90% | 516 |
| Xenopus laevis | African Clawed Frog | 371.2 | NP_001087058.1 | 78.70% | 489 |
| Danio rerio | Zebra Fish | 400.1 | NP_001004551.1 | 76.20% | 486 |
| Latimeria chalumnae | Coelacanth | 414.9 | XP_00599466.3 | 83.50% | 478 |
| Saccoglossus kowalevskii | Acorn Worm | 661.2 | XP_006822108 | 50.50% | 496 |
| Pediculus humanus corporis | Human Body Lice | 782.7 | XP_002424359 | 46.10% | 447 |
| Acyrthosiphon pison | Aphid | 782.7 | NP_001162147 | 43.50% | 449 |
| Caenorhabditis elegans | Roundworm | 937.5 | NP_497788.1 | 35.10% | 442 |
References
- ↑ "AceView". NCBI. http://www.ncbi.nlm.gov/ieb/research/acembly/index.html. Retrieved 1 March 2014.[yes|permanent dead link|dead link}}]
- ↑ "CCDC47 coiled-coil domain containing 47". NCBI. https://www.ncbi.nlm.nih.gov/gene/57003. Retrieved 3 March 2014.
- ↑ "SAPS Anaysis". SDSC Workbench. http://workbench.sdsc.edu. Retrieved 14 April 2014.
- ↑ "NCBI BLAST". National Center for Biotechnology Information. http://blast.ncbi.nlm.hin.gov/Blast. Retrieved 7 March 2014.[yes|permanent dead link|dead link}}]
- ↑ "Genecards". The Human Gene Compendium. https://www.genecards.org. Retrieved 7 March 2014.
- ↑ "Sulfinator". ExPASy. http://web.expasy.org/sulfinator. Retrieved 7 April 2014.
- ↑ "PHYRE 2 Protein Recognition Software". http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id-index. Retrieved 14 April 2014.
- ↑ "Coiled coil domains: stability, specificity, and biological implications". ChemBioChem 5 (2): 170–6. 2004. doi:10.1002/cbic.200300781. PMID 14760737.
- ↑ "El Dorado". Genomatix. http://www.genomatix.de/cgi-bin//eldorado. Retrieved 3 April 2014.[yes|permanent dead link|dead link}}]
- ↑ "Protein One". Transcription Factors. http://www.proteinone.com/products/proteins/transcription-factors/. Retrieved 29 March 2014.
- ↑ "Protein Spotlight, The PAR b ZIP Family". http://web.expasy.org/spotlight/snapshots/002/. Retrieved March 28, 2014.
- ↑ "The mfold Web Server". http://mfold.rna.albany.edu/?q-mfold. Retrieved 3 April 2014.
- ↑ "DAS-TM Filter Server". ExPASy. http://mendel.imp.ac.at/sat/DAS/DAS.html. Retrieved 17 April 2014.
- ↑ "NetPhos Server 2.0". ExPASy. http://www.cbs.dtu.dk/services/NetPhos. Retrieved 20 April 2014.
- ↑ Viguerie, Nathalie; Picard, Flora; Hul, Gabby; Roussel, Balbine; Barbe, Pierre; Iacovoni, Jason S.; Valle, Carine; Langin, Dominique et al. (2012). "Multiple effects of a short-term dexamethasone treatment in human skeletal muscle and adipose tissue". Physiological Genomics 44 (2): 141–151. doi:10.1152/physiolgenomics.00032.2011. ISSN 1094-8341. PMID 22108209.
- ↑ "Sulfinator". ExPASy. http://web.expasy.org/sulfinator. Retrieved 20 April 2014.
- ↑ "SumoPLOT". ExPASy. http://www.abgent.com/sumplot. Retrieved 20 April 2014.[yes|permanent dead link|dead link}}]
- ↑ "GEO Profiles". NCBI. https://www.ncbi.nlm.nih.gov/geo. Retrieved 20 March 2014.
- ↑ "Time Tree: The Timescale of Life". http://www.timetree.org/. Retrieved 13 March 2014.
- ↑ "BLAST". NCBI. https://www.ncbi.nlm.nih.gov/BLAST/%7Cpublisher=NCBI. Retrieved 13 March 2014.
External links
- Human CCDC47 genome location and CCDC47 gene details page in the UCSC Genome Browser.
 |
