Biology:Carboxysome
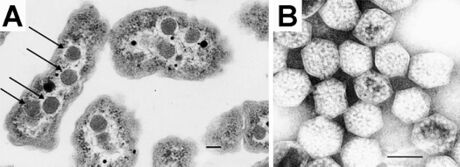
Carboxysomes are bacterial microcompartments (BMCs) consisting of polyhedral protein shells filled with the enzymes ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO)—the predominant enzyme in carbon fixation and the rate limiting enzyme in the Calvin cycle—and carbonic anhydrase.[2]
Carboxysomes are thought to have evolved as a consequence of the increase in oxygen concentration in the ancient atmosphere; this is because oxygen is a competing substrate to carbon dioxide in the RuBisCO reaction.[3] To overcome the inefficiency of RuBisCO, carboxysomes concentrate carbon dioxide inside the shell by means of co-localized carbonic anhydrase activity, which produces carbon dioxide from the bicarbonate that diffuses into the carboxysome. The resulting concentration of carbon dioxide near RuBisCO decreases the proportion of ribulose-1,5-bisphosphate oxygenation and thereby avoids costly photorespiratory reactions. The surrounding shell provides a barrier to carbon dioxide loss, helping to increase its concentration around RuBisCO.[4][5][6]
Carboxysomes are an essential part of the broader metabolic network called the Carbon dioxide-Concentrating Mechanism (CCM), which functions in two parts:[7] (1) Membrane transporters concentrate inorganic carbon (Ci) in the cell cytosol which is devoid of carbonic anhydrases. Carbon is primarily stored in the form of HCO3- which cannot re-cross the lipid membrane, as opposed to neutral CO2 which can easily escape the cell. This stockpiles carbon in the cell, creating a disequilibrium between the intracellular and extracellular environments of about 30x the Ci concentration in water.[8] (2) Cytosolic HCO3- diffuses into the carboxysome, where carboxysomal carbonic anhydrases dehydrate it back to CO2 in the vicinity of Rubisco, allowing Rubisco to operate at its maximal rate.
Carboxysomes are the best studied example of bacterial microcompartments, the term for functionally diverse organelles that are alike in having a protein shell.[9][10]
Discovery
Polyhedral bodies were discovered by transmission electron microscopy in the cyanobacterium Phormidium uncinatum in 1956.[11] These were later observed in other cyanobacteria[12] and in some chemotrophic bacteria that fix carbon dioxide—many of them are sulfur oxidizers or nitrogen fixers (for example, Halothiobacillus, Acidithiobacillus, Nitrobacter and Nitrococcus; all belonging to Pseudomonadota).[2][13] The polyhedral bodies were first purified from Thiobacillus neapolitanus (now Halothiobacillus neapolitanus) in 1973 and shown to contain RuBisCO, held within a rigid outer covering.[14] The authors proposed that since these appeared to be organelles involved in carbon dioxide fixation, they should be called carboxysomes.[14]
Architecture

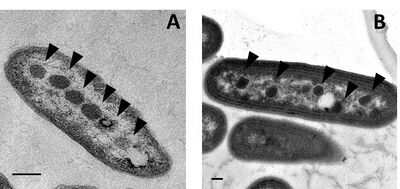
Structurally, carboxysomes are icosahedral, or quasi-icosahedral. Electron cryo-tomography studies[15][16][17] have confirmed the approximately icosahedral geometry of the carboxysome, and have imaged Rubisco proteins inside arranged in a few concentric layers or fibril-like structures.[15][17][18] The non-icosahedral faceted shapes of some carboxysomes can naturally be explained within the elastic theory of heterogeneous thin shells.[19]
Shell proteins
The carboxysome has an outer shell composed of a few thousand protein subunits, with hexameric shell proteins populating the faces and pentameric shell proteins placed at the 12 icosahedral vertices.[20] Proteins known to form the shell have been structurally characterized by X-ray crystallography. The proteins that constitute the majority of the shell form cyclical hexamers or pseudo-hexamers and belong to the BMC protein family.[21] Small pores perforate many different types of BMC-H hexamers, and may serve as the route for diffusion of small substrates (e.g. bicarbonate) and products (3-phosphoglycerate) into and out of the carboxysome. Positively charged amino acids in the pores presumably help promote the diffusion of the negatively charged substrates and products.[21] Other minor structural components of the shell that have been characterized include pentameric proteins (BMC-P proteins) which occupy the vertices of the icosahedral shell.[22] A third building block of the carboxysome shell is a protein composed of two BMC domains in tandem (BMC-T proteins). Structurally, these are known to form trimers which are pseudohexameric.[23][24] Some members of the BMC-T protein family stack in a face-to-face fashion and form tiny cages, notably both types of carboxysomes (alpha and beta, see below) contain these stacking trimers.[23][24] Based on crystal structures, these protein cages have relatively large gated pores on both sides, and it has been proposed that the opening and closing of the pore could be controlled in a manner similar to an air-lock. Such an air-lock, in contrast to BMC-H proteins with constitutively open pores, has been suggested to serve as a route for larger substrates (ribulose-1,5-bisphosphate) and products (3-phosphoglycerate) that must cross the shell.[23][24]
Production of empty carboxysome shells in E. coli enabled the first visualization of the carboxysome shell by cryo-electron microscopy.[25]
A number of viral capsids are also icosahedral, composed of hexameric and pentameric proteins, but currently there is no evidence suggesting any evolutionary relationship between the carboxysome shell and viral capsids.[26]
Scaffold proteins
All carboxysomes contain scaffold proteins that nucleate carboxysome components together during the assembly process. These scaffold proteins are required for carboxysome assembly; without them, carboxysomes do not form.[27] The α-carboxysomal scaffold protein is called CsoS2, and the β-carboxysomal scaffold protein is called CcmM. Though CsoS2 and CcmM have related functions, they have no evolutionary or sequence similarity. Both proteins bind to Rubisco, thereby ensuring that Rubisco gets packaged during carboxysome biogenesis.[28][29] Remarkably, both proteins bind to Rubisco at a binding site that bridges two large subunits while maintaining contact with the small subunit, ensuring that only the 16-subunit Rubisco holoenzyme is encapsulated. Both CsoS2 and CcmM have repetitive domain structures giving them multi-valent modes of binding. CcmM has three small-subutnit-like (SSUL) domains that bind to Rubisco,[29] and CsoS2 has four N-terminal domain (NTD) repeats that bind Rubisco,[28] making it possible for each single scaffold protein to bind up to 3-4 Rubiscos at a time. CsoS2 has also been shown to bind to shell proteins via its 7 Middle Region (MR) repeats and C-terminal domain (CTD).[27][30] In α-carboxysomes, the CsoS2 MR repeats have been shown to define the size of the carboxysome.[31]
Two types of carboxysomes
There are two types of carboxysomes. Although they may seem similar in appearance, they differ in their protein composition, including the form of RuBisCO they enclose.[32][33][34][35] Furthermore, studies have revealed fundamental differences in their gene organization and possibly their assembly pathway. Based on bioinformatic studies of shell proteins, it appears that the two types of carboxysomes evolved independently.[36][35]
Alpha-carboxysomes
Alpha-carboxysomes (aka α-carboxysomes) are also referred as the cso type of carboxysome. They contain Form IA RuBisCO; they are found in alpha-cyanobacteria, some nitrifying bacteria, some sulfur-oxidizing bacteria (for example, Halothiobacillus neapolitanus), and some purple bacteria; these are all classified as Pseudomonadota). The alpha-carboxysome was the first bacterial microcompartment to be purified and characterized.[37][38] Electron microscopy studies on purified alpha-carboxysomes or cell sections containing alpha-carboxysomes revealed that they are typically 100-160 nm in diameter.[39] Common building blocks for the shell of alpha-carboxysomes are called CsoS1A/B/C (BMC-H), CsoS4A/B (BMC-P), and CsoS1D (BMC-T). CsoS4A/B were the first BMC-P proteins to be experimentally demonstrated as minor components of the BMC shell[4] (only 12 pentamers are required to cap the vertices of an icosahedron). CsoS1D is the first BMC-T which has been structurally characterized; it is also the first example of dimerization of two BMC building blocks in a face-to-face fashion to create a tiny cage. The CsoS1D cage has a gated pore at both ends, which is proposed to facilitate the transfer of large metabolites across the shell.[24] In addition to the specific form of RuBisCO, other encapsulated proteins distinguish alpha-carboxysomes from beta-carboxysomes such as scaffold protein CsoS2 and carbonic anhydrase CsoSCA. CsoS2 is an intrinsically disordered protein with an essential role in alpha-carboxysome assembly. It has a very high pI and a unique primary structure with three domains: an N-terminal, a middle- and a C-terminal domain.[27][40] Repetitive motifs can be identified in all three regions; the N-terminal domain repeats bind to Rubisco,[28] the middle region domains bind to shell proteins,[30] and the c-terminal domain repeats also bind to shell proteins.[41][42][43] CsoSCA is a beta-carbonic anhydrase that binds to Rubisco.[5][44][45] Studies in Halothiobacillus neapolitanus have shown that empty shells of normal shape and composition are assembled in carboxysomal RuBisCO-lacking mutants, suggesting that alpha-carboxysome shell biogenesis and enzyme sequestration are two independent, but functionally linked processes.[46] Intriguingly, carboxysomes of Halothiobacillus neapolitanus have been found to accommodate chimeric and heterologous species of RuBisCO. It is the large subunit of RuBisCO which determines whether the enzyme is sequestered into carboxysomes.[46]
Beta-carboxysomes
Beta-carboxysomes (aka β-carboxysomes) are found in cyanobacteria.[47]
The signature proteins of the beta-carboxysome are Form IB RuBisCO and a gamma carbonic anhydrase homolog.[9] Beta-carboxysomes are typically larger than alpha-carboxysomes: the observed diameters vary from 200 to 400 nm.[27] The structural proteins that are essential for beta-carboxysome formation are encoded in the conserved carboxysome locus[10] known as the ccm locus. The ccm locus includes genes for core proteins CcmM and CcmN and the shell proteins CcmK (a BMC-H protein), CcmL (a BMC-P protein) and CcmO (a BMC-T protein).
A full length CcmM protein consists of a gamma-carbonic anhydrase domain and three to five RubisCO small subunit-like domains (SSLDs) on its C-terminus.[48] The ccmM gene contains an internal translation site that produces a short form of CcmM which only consists of SSLDs; both long and short forms of CcmM are required for beta-carboxysome assembly.[49] CcmN contains multiple hexapeptide-repeat domains on its N-terminus and a short α-helical encapsulation peptide on the C-terminus.[50]
Other structural components of beta-carboxysomes are encoded outside of the ccm locus. CcmP is a BMC-T protein that is absolutely conserved among organisms that form beta-carboxysomes. Two CcmP pseudohexamers stack to form a nanocompartment—an example of an air-lock forming protein.[23] Likewise, in some cyanobacterial strains the beta-carboxysomes contain a beta-carbonic anhydrase that is not encoded in the ccm locus.[51]
Shell proteins of beta carboxysomes are relatively diverse[47] compared to their counterparts in the alpha carboxysomes, and this has been proposed to reflect variable permeability requirements of beta carboxysomes, which are found in cyanobacteria that occupy ecophysiologically dynamic environments.[52]
The beta-carboxysome assembles from the inside out. First an enzymatic core forms that is subsequently encapsulated by the protein shell.[53] Carboxysome assembly occurs through a series of protein-protein interactions: the enzyme RuBisCO and the two isoforms (full length and short form) of the CcmM protein interact by means of the SSLDs; in strains containing CcaA the beta-carbonic anhydrase is brought into the carboxysome core by interaction with the N-terminus of the full length CcmM.[54][55] Once the procarboxysome (the carboxysome core) is formed, the N-terminus of the adapter protein CcmN interacts with the N-terminus of CcmM, while the C-terminus of CcmN recruits the shell proteins CcmK (BMC-H) and CcmO (BMC-T), utilizing a 15-20 amino acids long peptide.[50] This encapsulation peptide forms an amphipathic a-helix that interacts with the shell components and its role is essential, given that in its absence, carboxysomes cannot be formed.[50][35] The final step is the addition of the vertices formed by the BMC-P protein CcmL, which then cap the enzymatic core and facets.[53] Elucidation of the assembly pathway of beta carboxysomes enabled the design of a single synthetic protein that replaced four other proteins in carboxysome assembly.[56]
Potential uses of the carboxysome in biotechnology
As is the case with other BMCs, the carboxysome is attracting significant attention by researchers for applications in plant synthetic biology.[57][32][58] The transfer of a genetic module coding for an alpha-carboxysome has been shown to produce carboxysome-like structures in E. coli.[59] Bioengineering of carboxysome shells has been shown to be feasible, and beta-carboxysomes constructed with chimeric proteins or with chimeric shells have been reported.[60] The introduction of carboxysomes into plant chloroplasts as part of a CO
2 concentrating mechanism [61][62] such as that found in cyanobacteria is predicted to significantly improve net CO
2 fixation and yield.[63][64] Expression of beta-carboxysomal shell proteins [65] and Form IB Rubisco-CcmM complexes in tobacco chloroplasts has been achieved,[66] but did not result in compartments containing RuBisCO. A further advance has been the construction of minimal alpha-carboxysomes containing Form IA Rubisco and the CsoS1A and CsoS2 proteins from the cyanobacterium Cyanobium PCC7001 in tobacco chloroplasts.[67] As yet, identifiably functional carboxysomes have not been constructed in plant chloroplasts. Improvement of photosynthesis in plants using this approach is ultimately dependent on the operation of transporter proteins in the chloroplast inner envelope membrane to help generate a high concentration of bicarbonate inside the chloroplast.[68]
Potential applications of carboxysomes (list format):
- Engineer the carbon-concentrating mechanism (CCM) and carboxysomes into industrially relevant microbes, potentially converting heterotrophic organisms into mixotrophs or autotrophs that capture CO2 while producing high value products.[69]
- Engineer the carbon-concentrating mechanism (CCM) and carboxysomes into plants for increased CO2 capture and enhanced growth.
- Engineer faster Rubiscos. The fastest form I prokaryotic Rubiscos are mostly found in α-carboxysomes.[70]
- Engineer a minimal carboxysome gene set (Rubisco, carbonic anhydrase, scaffold protein, hexameric shell, pentameric shell) to facilitate facile engineering into alternative host organisms.
- Design in vitro carboxysomes for cell-free CO2 fixation.
- Engineer carboxysomes to have alternative metabolisms.[42][71]
Carboxysome reviews (by year)
Carboxysome research expands every year. Published reviews chart the rapid pace of discovery across the broad field of "carboxysomics".
| First Author | Title | Year | Link |
|---|---|---|---|
| Shively et al. | Inclusion bodies of prokaryotes | 1974 | [1] |
| Badger and Price | The CO2 concentrating mechanism in cyanobacteria and microalgae | 1992 | [2] |
| Giordano et al. | CO2 CONCENTRATING MECHANISMS IN ALGAE: Mechanisms, Environmental Modulation, and Evolution | 2005 | [3] |
| Heinhorst et al. | Carboxysomes and Carboxysome-like Inclusions | 2006 | [4] |
| Espie et al. | Carboxysomes: cyanobacterial RubisCO comes in small packages | 2011 | [5] |
| Kinney et al. | Comparative analysis of carboxysome shell proteins | 2011 | [6] |
| Moroney et al. | Photorespiration and carbon concentrating mechanisms: two adaptations to high O2, low CO2 conditions | 2013 | [7] |
| Rae et al. | Functions, Compositions, and Evolution of the Two Types of Carboxysomes: Polyhedral Microcompartments That Facilitate CO2 Fixation in Cyanobacteria and Some Proteobacteria | 2013 | [8] |
| Hanson et al. | Towards engineering carboxysomes into C3 plants | 2016 | [9] |
| Kerfeld and Melnicki | Assembly, function and evolution of cyanobacterial carboxysomes | 2016 | [10] |
| Rae et al. | Progress and challenges of engineering a biophysical CO2-concentrating mechanism into higher plants | 2017 | [11] |
| Turmo et al. | Carboxysomes: metabolic modules for CO2 fixation | 2017 | [12] |
| Hennacy and Jonikas | Prospects for Engineering Biophysical CO2 Concentrating Mechanisms into Land Plants to Enhance Yields | 2020 | [13] |
| Borden and Savage | New discoveries expand possibilities for carboxysome engineering | 2021 | [14] |
| Huffine et al. | Computational modeling and evolutionary implications of biochemical reactions in bacterial microcompartments | 2021 | [15] |
| Liu | Advances in the bacterial organelles for CO2 fixation | 2021 | [16] |
| Liu et al. | Protein stoichiometry, structural plasticity and regulation of bacterial microcompartments | 2021 | [17] |
See also
References
- ↑ "Structural analysis of CsoS1A and the protein shell of the Halothiobacillus neapolitanus carboxysome". PLOS Biology 5 (6): e144. June 2007. doi:10.1371/journal.pbio.0050144. PMID 17518518.
- ↑ 2.0 2.1 "Protein-based organelles in bacteria: carboxysomes and related microcompartments". Nature Reviews. Microbiology 6 (9): 681–691. September 2008. doi:10.1038/nrmicro1913. PMID 18679172.
- ↑ "CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution". Journal of Experimental Botany 54 (383): 609–622. February 2003. doi:10.1093/jxb/erg076. PMID 12554704.
- ↑ 4.0 4.1 "The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to function as a CO2 leakage barrier". PLOS ONE 4 (10): e7521. October 2009. doi:10.1371/journal.pone.0007521. PMID 19844578. Bibcode: 2009PLoSO...4.7521C.
- ↑ 5.0 5.1 "CO2 fixation kinetics of Halothiobacillus neapolitanus mutant carboxysomes lacking carbonic anhydrase suggest the shell acts as a diffusional barrier for CO2". The Journal of Biological Chemistry 283 (16): 10377–10384. April 2008. doi:10.1074/jbc.M709285200. PMID 18258595.
- ↑ "pH determines the energetic efficiency of the cyanobacterial CO2 concentrating mechanism". Proceedings of the National Academy of Sciences of the United States of America 113 (36): E5354–E5362. September 2016. doi:10.1073/pnas.1525145113. PMID 27551079. Bibcode: 2016PNAS..113E5354M.
- ↑ "Evolution and diversity of CO2 concentrating mechanisms in cyanobacteria". Functional Plant Biology 29 (3): 161–173. April 2002. doi:10.1071/PP01213. PMID 32689463.
- ↑ "Inorganic carbon transporters of the cyanobacterial CO2 concentrating mechanism". Photosynthesis Research 109 (1–3): 47–57. September 2011. doi:10.1007/s11120-010-9608-y. PMID 21359551.
- ↑ 9.0 9.1 "Bacterial microcompartments and the modular construction of microbial metabolism". Trends in Microbiology 23 (1): 22–34. January 2015. doi:10.1016/j.tim.2014.10.003. PMID 25455419.
- ↑ 10.0 10.1 "A taxonomy of bacterial microcompartment loci constructed by a novel scoring method". PLOS Computational Biology 10 (10): e1003898. October 2014. doi:10.1371/journal.pcbi.1003898. PMID 25340524. Bibcode: 2014PLSCB..10E3898A.
- ↑ "[Cytology of Cyanophycea. II. Centroplasm and granular inclusions of Phormidium uncinatum]". Archiv für Mikrobiologie 24 (2): 147–162. 1956. doi:10.1007/BF00408629. PMID 13327992.
- ↑ "Ultrastructure of blue-green algae". Journal of Bacteriology 97 (3): 1486–1493. March 1969. doi:10.1128/JB.97.3.1486-1493.1969. PMID 5776533.
- ↑ "Inclusion bodies of prokaryotes". Annual Review of Microbiology 28: 167–187. 1974. doi:10.1146/annurev.mi.28.100174.001123. PMID 4372937.
- ↑ 14.0 14.1 "Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus". Science 182 (4112): 584–586. November 1973. doi:10.1126/science.182.4112.584. PMID 4355679. Bibcode: 1973Sci...182..584S.
- ↑ 15.0 15.1 "The structure of isolated Synechococcus strain WH8102 carboxysomes as revealed by electron cryotomography". Journal of Molecular Biology 372 (3): 764–773. September 2007. doi:10.1016/j.jmb.2007.06.059. PMID 17669419.
- ↑ "Organization, structure, and assembly of alpha-carboxysomes determined by electron cryotomography of intact cells". Journal of Molecular Biology 396 (1): 105–117. February 2010. doi:10.1016/j.jmb.2009.11.019. PMID 19925807.
- ↑ 17.0 17.1 "Structure of Halothiobacillus neapolitanus carboxysomes by cryo-electron tomography". Journal of Molecular Biology 364 (3): 526–535. December 2006. doi:10.1016/j.jmb.2006.09.024. PMID 17028023.
- ↑ "Rubisco forms a lattice inside alpha-carboxysomes". Nature Communications 13 (1): 4863. August 2022. doi:10.1038/s41467-022-32584-7. PMID 35982043. Bibcode: 2022NatCo..13.4863M.
- ↑ "Platonic and Archimedean geometries in multicomponent elastic membranes". Proceedings of the National Academy of Sciences of the United States of America 108 (11): 4292–4296. March 2011. doi:10.1073/pnas.1012872108. PMID 21368184.
- ↑ "Comparative analysis of carboxysome shell proteins". Photosynthesis Research 109 (1–3): 21–32. September 2011. doi:10.1007/s11120-011-9624-6. PMID 21279737.
- ↑ 21.0 21.1 "Protein structures forming the shell of primitive bacterial organelles". Science 309 (5736): 936–938. August 2005. doi:10.1126/science.1113397. PMID 16081736. Bibcode: 2005Sci...309..936K.
- ↑ "Atomic-level models of the bacterial carboxysome shell". Science 319 (5866): 1083–1086. February 2008. doi:10.1126/science.1151458. PMID 18292340. Bibcode: 2008Sci...319.1083T.
- ↑ 23.0 23.1 23.2 23.3 "The structure of CcmP, a tandem bacterial microcompartment domain protein from the β-carboxysome, forms a subcompartment within a microcompartment". The Journal of Biological Chemistry 288 (22): 16055–16063. May 2013. doi:10.1074/jbc.M113.456897. PMID 23572529.
- ↑ 24.0 24.1 24.2 24.3 "Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport". Journal of Molecular Biology 392 (2): 319–333. September 2009. doi:10.1016/j.jmb.2009.03.056. PMID 19328811.
- ↑ "Structure of a Synthetic β-Carboxysome Shell". Plant Physiology 181 (3): 1050–1058. November 2019. doi:10.2210/pdb6owg/pdbx. PMID 31501298.
- ↑ "Cellular origin of the viral capsid-like bacterial microcompartments". Biology Direct 12 (1): 25. November 2017. doi:10.1186/s13062-017-0197-y. PMID 29132422.
- ↑ 27.0 27.1 27.2 27.3 "Advances in Understanding Carboxysome Assembly in Prochlorococcus and Synechococcus Implicate CsoS2 as a Critical Component". Life 5 (2): 1141–1171. March 2015. doi:10.3390/life5021141. PMID 25826651. Bibcode: 2015Life....5.1141C.
- ↑ 28.0 28.1 28.2 "Multivalent interactions between CsoS2 and Rubisco mediate α-carboxysome formation". Nature Structural & Molecular Biology 27 (3): 281–287. March 2020. doi:10.1038/s41594-020-0387-7. PMID 32123388.
- ↑ 29.0 29.1 "Rubisco condensate formation by CcmM in β-carboxysome biogenesis". Nature 566 (7742): 131–135. February 2019. doi:10.1038/s41586-019-0880-5. PMID 30675061. Bibcode: 2019Natur.566..131W.
- ↑ 30.0 30.1 "Conserved and repetitive motifs in an intrinsically disordered protein drive α-carboxysome assembly". bioRxiv (Cold Spring Harbor Laboratory (CSHL)). 2023-07-08. doi:10.1101/2023.07.08.548221.
- ↑ "α-carboxysome size is controlled by the disordered scaffold protein CsoS2" (in en). bioRxiv (Cold Spring Harbor Laboratory (CSHL)). 2023-07-08. doi:10.1101/2023.07.07.548173.
- ↑ 32.0 32.1 "Cyanobacterial-based approaches to improving photosynthesis in plants". Journal of Experimental Botany 64 (3): 787–798. January 2013. doi:10.1093/jxb/ers294. PMID 23095996.
- ↑ "Functions, compositions, and evolution of the two types of carboxysomes: polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some proteobacteria". Microbiology and Molecular Biology Reviews 77 (3): 357–379. September 2013. doi:10.1128/MMBR.00061-12. PMID 24006469.
- ↑ "Carboxysomes: metabolic modules for CO2 fixation". FEMS Microbiology Letters 364 (18). October 2017. doi:10.1093/femsle/fnx176. PMID 28934381.
- ↑ 35.0 35.1 35.2 "Assembly, function and evolution of cyanobacterial carboxysomes". Current Opinion in Plant Biology 31: 66–75. June 2016. doi:10.1016/j.pbi.2016.03.009. PMID 27060669.
- ↑ "Evolutionary relationships among shell proteins of carboxysomes and metabolosomes". Current Opinion in Microbiology 63: 1–9. October 2021. doi:10.1016/j.mib.2021.05.011. PMID 34098411.
- ↑ "Icosahedral inclusions (carboxysomes) of Nitrobacter agilis". Journal of Bacteriology 132 (2): 673–675. November 1977. doi:10.1128/JB.132.2.673-675.1977. PMID 199579.
- ↑ "Characterization of a homogenous preparation of carboxysomes from Thiobacillus neapolitanus". Archives of Microbiology 134 (1): 52–59. 1983. doi:10.1007/BF00429407. ISSN 0302-8933.
- ↑ "Carboxysomes and Their Structural Organization in Prokaryotes". Nanomicrobiology. 2014. pp. 75–101. doi:10.1007/978-1-4939-1667-2_4. ISBN 978-1-4939-1666-5.
- ↑ "New discoveries expand possibilities for carboxysome engineering". Current Opinion in Microbiology 61: 58–66. June 2021. doi:10.1016/j.mib.2021.03.002. PMID 33798818.
- ↑ "Structure of a Minimal α-Carboxysome-Derived Shell and Its Utility in Enzyme Stabilization". Biomacromolecules 22 (10): 4095–4109. October 2021. doi:10.1021/acs.biomac.1c00533. PMID 34384019.
- ↑ 42.0 42.1 "Reprogramming bacterial protein organelles as a nanoreactor for hydrogen production". Nature Communications 11 (1): 5448. October 2020. doi:10.1038/s41467-020-19280-0. PMID 33116131. Bibcode: 2020NatCo..11.5448L.
- ↑ "Intrinsically disordered CsoS2 acts as a general molecular thread for α-carboxysome shell assembly.". bioRxiv (Cold Spring Harbor Laboratory (CSHL)). 2023. doi:10.1101/2023.06.24.546370.
- ↑ "The structure of beta-carbonic anhydrase from the carboxysomal shell reveals a distinct subclass with one active site for the price of two". The Journal of Biological Chemistry 281 (11): 7546–7555. March 2006. doi:10.1074/jbc.M510464200. PMID 16407248.
- ↑ "Discovery of a carbonic anhydrase-Rubisco complex within the alpha-carboxysome.". bioRxiv (Cold Spring Harbor Laboratory (CSHL)). 2023. doi:10.1101/2021.11.05.467472.
- ↑ 46.0 46.1 "Halothiobacillus neapolitanus carboxysomes sequester heterologous and chimeric RubisCO species". PLOS ONE 3 (10): e3570. 2008. doi:10.1371/journal.pone.0003570. PMID 18974784. Bibcode: 2008PLoSO...3.3570M.
- ↑ 47.0 47.1 "β-Carboxysome bioinformatics: identification and evolution of new bacterial microcompartment protein gene classes and core locus constraints". Journal of Experimental Botany 68 (14): 3841–3855. June 2017. doi:10.1093/jxb/erx115. PMID 28419380.
- ↑ "Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple Rubisco complexes with carboxysomal proteins CcmM and CcaA". The Journal of Biological Chemistry 282 (40): 29323–29335. October 2007. doi:10.1074/jbc.M703896200. PMID 17675289.
- ↑ "Functional cyanobacterial beta-carboxysomes have an absolute requirement for both long and short forms of the CcmM protein". Plant Physiology 153 (1): 285–293. May 2010. doi:10.1104/pp.110.154948. PMID 20304968.
- ↑ 50.0 50.1 50.2 "Elucidating essential role of conserved carboxysomal protein CcmN reveals common feature of bacterial microcompartment assembly". The Journal of Biological Chemistry 287 (21): 17729–17736. May 2012. doi:10.1074/jbc.M112.355305. PMID 22461622.
- ↑ "Carboxysomal carbonic anhydrases: Structure and role in microbial CO2 fixation". Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 1804 (2): 382–392. February 2010. doi:10.1016/j.bbapap.2009.09.026. PMID 19818881. https://digital.library.unt.edu/ark:/67531/metadc840980/.
- ↑ "Heterohexamers Formed by CcmK3 and CcmK4 Increase the Complexity of Beta Carboxysome Shells". Plant Physiology 179 (1): 156–167. January 2019. doi:10.1104/pp.18.01190. PMID 30389783.
- ↑ 53.0 53.1 "Biogenesis of a bacterial organelle: the carboxysome assembly pathway". Cell 155 (5): 1131–1140. November 2013. doi:10.1016/j.cell.2013.10.044. PMID 24267892.
- ↑ "A multiprotein bicarbonate dehydration complex essential to carboxysome function in cyanobacteria". Journal of Bacteriology 190 (3): 936–945. February 2008. doi:10.1128/JB.01283-07. PMID 17993516.
- ↑ "Over-expression of the β-carboxysomal CcmM protein in Synechococcus PCC7942 reveals a tight co-regulation of carboxysomal carbonic anhydrase (CcaA) and M58 content". Photosynthesis Research 109 (1–3): 33–45. September 2011. doi:10.1007/s11120-011-9659-8. PMID 21597987.
- ↑ "Streamlined Construction of the Cyanobacterial CO2-Fixing Organelle via Protein Domain Fusions for Use in Plant Synthetic Biology". The Plant Cell 27 (9): 2637–2644. September 2015. doi:10.1105/tpc.15.00329. PMID 26320224.
- ↑ "Plug-and-play for improving primary productivity". American Journal of Botany 102 (12): 1949–1950. December 2015. doi:10.3732/ajb.1500409. PMID 26656128.
- ↑ "Bacterial microcompartments as metabolic modules for plant synthetic biology". The Plant Journal 87 (1): 66–75. July 2016. doi:10.1111/tpj.13166. PMID 26991644.
- ↑ "Modularity of a carbon-fixing protein organelle". Proceedings of the National Academy of Sciences of the United States of America 109 (2): 478–483. January 2012. doi:10.1073/pnas.1108557109. PMID 22184212.
- ↑ "Engineering bacterial microcompartment shells: chimeric shell proteins and chimeric carboxysome shells". ACS Synthetic Biology 4 (4): 444–453. April 2015. doi:10.1021/sb500226j. PMID 25117559.
- ↑ "Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants". Journal of Experimental Botany 59 (7): 1441–1461. 2008. doi:10.1093/jxb/erm112. PMID 17578868.
- ↑ "The cyanobacterial CCM as a source of genes for improving photosynthetic CO2 fixation in crop species". Journal of Experimental Botany 64 (3): 753–768. January 2013. doi:10.1093/jxb/ers257. PMID 23028015.
- ↑ "Can the cyanobacterial carbon-concentrating mechanism increase photosynthesis in crop species? A theoretical analysis". Plant Physiology 164 (4): 2247–2261. April 2014. doi:10.1104/pp.113.232611. PMID 24550242.
- ↑ "Can increased leaf photosynthesis be converted into higher crop mass production? A simulation study for rice using the crop model GECROS". Journal of Experimental Botany 68 (9): 2345–2360. April 2017. doi:10.1093/jxb/erx085. PMID 28379522.
- ↑ "β-Carboxysomal proteins assemble into highly organized structures in Nicotiana chloroplasts". The Plant Journal 79 (1): 1–12. July 2014. doi:10.1111/tpj.12536. PMID 24810513.
- ↑ "A faster Rubisco with potential to increase photosynthesis in crops". Nature 513 (7519): 547–550. September 2014. doi:10.1038/nature13776. PMID 25231869. Bibcode: 2014Natur.513..547L.
- ↑ "Carboxysome encapsulation of the CO2-fixing enzyme Rubisco in tobacco chloroplasts". Nature Communications 9 (1): 3570. September 2018. doi:10.1038/s41467-018-06044-0. PMID 30177711. Bibcode: 2018NatCo...9.3570L.
- ↑ "Progress and challenges of engineering a biophysical CO2-concentrating mechanism into higher plants". Journal of Experimental Botany 68 (14): 3717–3737. June 2017. doi:10.1093/jxb/erx133. PMID 28444330..
- ↑ "Functional reconstitution of a bacterial CO2 concentrating mechanism in Escherichia coli". eLife 9. October 2020. doi:10.7554/eLife.59882. PMID 33084575.
- ↑ "Systematic exploration of prokaryotic form I rubisco maximal carboxylation rates.". bioRxiv (Cold Spring Harbor Laboratory (CSHL)). 2023-07-27. doi:10.1101/2023.07.27.550689.
- ↑ "Synthetic engineering of a new biocatalyst encapsulating [NiFe-hydrogenases for enhanced hydrogen production"]. Journal of Materials Chemistry B 11 (12): 2684–2692. March 2023. doi:10.1039/D2TB02781J. PMID 36883480.
External links
- Mysterious Bacterial Microcompartments Revealed By Biochemists
- Not so simple after all. A renaissance of research into prokaryotic evolution and cell structure
 |
