Biology:Enhancer-FACS-seq
Enhancer-FACS-seq (eFS),[1][2] developed by the Bulyk lab[3] at Brigham and Women’s Hospital and Harvard Medical School,[4] is a highly parallel enhancer assay that aims for the identification of active, tissue-specific transcriptional enhancers, in the context of whole Drosophila melanogaster embryos. This technology replaces the use of microscopy to screen for tissue-specific enhancers with fluorescence activated cell sorting (FACS) of dissociated cells from whole embryos, combined with identification by high-throughput Illumina sequencing.
Introduction
Transcriptional regulation
In metazoans, in order to respond to environmental stress, differentiate properly, and progress normally through the cell cycle, a eukaryotic cell needs a specific and coordinated gene expression program, which involves the highly regulated transcription of thousands of genes.[5] This gene regulation is in large part controlled, in a tissue-specific manner, by the binding of transcription factors to noncoding genomic regions referred to as cis-regulatory modules (CRMs),[6] activating or repressing gene expression by modulating the structure of the chromatin and therefore having a positive or negative effect on transcription regulation. CRMs activating gene expression are often referred to as transcriptional enhancers, whereas those repressing gene expression are referred to as transcriptional silencers.
Enhancer detection in Drosophila melanogaster
Despite being a powerful model organism for biology and the study of transcriptional enhancers, the tissue specific activity of less than 5% of the estimated 50,000 transcriptional enhancers in Drosophila melanogaster have been discovered.[7][8] Over the past decade, the main method for detection of tissue- or cell-type specific activities of enhancers in Drosophila melanogaster was to test candidate enhancers by traditional reporter assays,[9][10] which are low-throughput and costly. Over the past few years, even though enhancer discovery has been improved and other parallel reporter assays have been developed,[11][12][13][14][15][16] none so far allowed the direct identification of enhancer activity in a genomic context in cell types of interest in a whole embryo.
Methodology
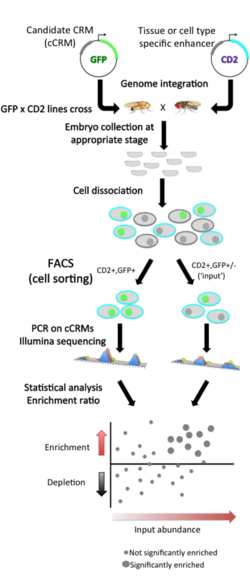
Each candidate CRM (cCRM) is cloned upstream of a reporter gene. Compared to traditional reporter assays, the main innovation is the use of fluorescence activated cell sorting (FACS) of dissociated cells, instead of microscopy, to screen for tissue-specific enhancers. This approach utilizes a two-marker system: in each embryo, one marker (here, the rat CD2 cell surface protein) is used to label cells of a specific tissue for being sorted by FACS, and the other marker (here, green fluorescent protein GFP) is used as a reporter of CRM activity.
Cells are sorted according to their tissue type and then by GFP fluorescence, and the cCRMs are recovered by PCR from double-positive sorted cells, and from total input cells. High-throughput sequencing of both populations then allows measuring the relative abundance of each cCRM in input and sorted populations; one can then assess the enrichment or depletion of each cCRM in double-positive cells versus input as a measure of activity in the CD2-positive cell type being tested.
Significant results
In the initial report on this method,[17] a library of ~500 cCRMs was drawn from a variety of genomic data sources (e.g., TF-bound regions, coactivator-bound regions, DNase I hypersensitive sites, and predictions from the Bulyk lab’s PhylCRM algorithm [18]) by PCR from genomic DNA, and then screened for activity in embryonic mesoderm and in specific mesodermal cell types. The results were validated by traditional reporter gene assay in Drosophila melanogaster embryos for 68 cCRMs tested by eFS. The specificity of eFS was excellent among significantly enriched cCRMs, while sensitivity was good where the majority of the CD2-positive cells express GFP. It was found that the known enhancer-associated chromatin marks H3K27ac, H3K4me1, and Pol II are significantly enriched among the enhancers found to be active in mesoderm.
Advantages and future applications
Advantages of eFS
- Highly parallel identification of active, tissue-specific transcriptional enhancers in whole embryos
- Candidate enhancers activity assayed in a genomic context
- High specificity of detected enhancers
Future applications
The eFS assay could be used to analyze other cell or tissue types. By assessing enrichment in GFP-expressing CD2-negative as well as CD2-positive cells, and by crossing a common pool of reporter transformant male flies to females expressing CD2 in different cell types, it is possible to assay specificity as well as activity. Accelerating the annotation of the regulatory genome in Drosophila should in principle generate the kind of large-scale regulatory interaction data that would allow exploring the network properties of transcriptional regulation.
References
- ↑ "Highly parallel enhancer assays". http://the_brain.bwh.harvard.edu/efs.html.
- ↑ Gisselbrecht SS (2013). "Highly parallel assays of tissue-specific enhancers in whole Drosophila embryos". Nat Methods 10 (8): 774–80. doi:10.1038/nmeth.2558. PMID 23852450. PMC 3733245. https://dash.harvard.edu/bitstream/handle/1/11879734/3733245.pdf?sequence=1.
- ↑ "Bulyk Lab". http://the_brain.bwh.harvard.edu/.
- ↑ "Bulyk Lab". http://the_brain.bwh.harvard.edu/.
- ↑ Lockhart, DJ; Winzeler, EA (2000). "Genomics, gene expression and DNA arrays.". Nature 405 (6788): 827–36. doi:10.1038/35015701. PMID 10866209.
- ↑ Bulyk ML (2003). "Computational prediction of transcription-factor binding site locations". Genome Biol. 5 (1): 201. doi:10.1186/gb-2003-5-1-201. PMID 14709165.
- ↑ Pfeiffer BD (2008). "Tools for neuroanatomy and neurogenetics in Drosophila.". Proc Natl Acad Sci U S A 105 (28): 9715–20. doi:10.1073/pnas.0803697105. PMID 18621688. Bibcode: 2008PNAS..105.9715P.
- ↑ Halfon MS (2007). "REDfly 2.0: an integrated database of cis-regulatory modules and transcription factor binding sites in Drosophila". Nucleic Acids Res 36 (Database issue): D594-8. doi:10.1093/nar/gkm876. PMID 18039705.
- ↑ Sandmann T (2007). "A core transcriptional network for early mesoderm development in Drosophila melanogaster". Genes Dev 21 (4): 436–49. doi:10.1101/gad.1509007. PMID 17322403.
- ↑ Zinzen RP (2009). "Combinatorial binding predicts spatio-temporal cis-regulatory activity". Nature 462 (7269): 65–70. doi:10.1038/nature08531. PMID 19890324. Bibcode: 2009Natur.462...65Z.
- ↑ Gertz J (2009). "Analysis of combinatorial cis-regulation in synthetic and genomic promoters". Nature 457 (7226): 215–8. doi:10.1038/nature07521. PMID 19029883. Bibcode: 2009Natur.457..215G.
- ↑ Sharon E (2012). "Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters". Nat Biotechnol 30 (6): 521–30. doi:10.1038/nbt.2205. PMID 22609971.
- ↑ Nam J (2010). "Functional cis-regulatory genomics for systems biology.". Proc. Natl. Acad. Sci. U.S.A. 107 (8): 3930–5. doi:10.1073/pnas.1000147107. PMID 20142491. Bibcode: 2010PNAS..107.3930N.
- ↑ Melnikov A (2012). "Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay". Nat Biotechnol 30 (3): 271–7. doi:10.1038/nbt.2137. PMID 22371084.
- ↑ Patwardhan RP (2012). "Massively parallel functional dissection of mammalian enhancers in vivo". Nat Biotechnol 30 (3): 265–70. doi:10.1038/nbt.2136. PMID 22371081.
- ↑ Arnold CD (2014). "Quantitative genome-wide enhancer activity maps for five Drosophila species show functional enhancer conservation and turnover during cis-regulatory evolution". Nat Genet 46 (7): 685–92. doi:10.1038/ng.3009. PMID 24908250.
- ↑ Gisselbrecht SS (2013). "Highly parallel assays of tissue-specific enhancers in whole Drosophila embryos". Nat Methods 10 (8): 774–80. doi:10.1038/nmeth.2558. PMID 23852450. PMC 3733245. https://dash.harvard.edu/bitstream/handle/1/11879734/3733245.pdf?sequence=1.
- ↑ "PhylCRM software package homepage". http://the_brain.bwh.harvard.edu/PhylCRM/index.html.
 |
