Biology:FAM120AOS
 Generic protein structure example |
FAM120AOS, or family with sequence similarity 120A opposite strand, codes for uncharacterized protein FAM120AOS, which currently has no known function.[1] The gene ontology describes the gene to be protein binding.[2] Overall, it appears that the thyroid and the placenta are the two tissues with the highest expression levels of FAM120AOS across a majority of datasets.
The microarray-assessed tissue expression pattern of multiple normal tissues for FAM120AOS in humans was found using GDS3834 data.[3] The three tissues in the 90th percentile and higher for FAM120AOS gene expression are as follows: the bladder, epididymis, and thyroid. The thyroid is in the 91st percentile, while the other two are in the 90th percentile. Since high thyroid expression was also seen across the RNA-seq data,[4][5][6][7] it appears that FAM120AOS expression may be important in the thyroid.
Gene
Common aliases
The common aliases for FAM120AOS are C9orf10OS, FLJ31534, LOC158293, and putative FAM120A opposite strand protein.[1]
Locus
There are two genomic locations for the gene, the first of which is chr9:93,431,441-93,453,601(GRCh38/hg38) with a length of 22,161 base pairs (bp), oriented on the minus strand of the chromosome.[8] The second genomic location for the gene is at chr9:96,208,776-96,215,874(GRCh37/hg19) with a length of 7,099 bp, also oriented on the minus strand of the chromosome.[8] The genes found upstream of FAM120AOS on chromosome 9 are FGD3, SUSD, C9orf89, WNK2, C9orf129, and NINJ1.[1] The genes found downstream from FAM120AOS on chromosome 9 are FAM120A and PHF2.[1]
Number of exons
The longest isoform of FAM120AOS in humans contains 3 exons.[2]
Span of gene
The mRNA transcript variant that encodes for human FAM120AOS isoform 1 is 5922 bp long and contains an upstream in-frame stop codon (taa) at 807-809 bp.[9]
Transcripts
There are 12 known isoforms of the human FAM120AOS gene.[2] The longest and most common transcript variant is isoform 1, which is 5922 bp in length.[2][10] Transcript variants 3-12 are all non-coding RNAs, meaning that they do not code for a protein.[2] The only isoforms that are protein-encoding are isoform 1 and 2 of the human FAM120AOS gene.[2]
Isoform 2 is 5008 bp in length and contains an alternate exon in the 5' UTR, is missing a portion of the 5' coding region, and initiates translation at an alternate start codon, in comparison to isoform 1.[2][11] The variant also has a shorter and more distinct N-terminus in comparison to isoform 1.[2]
Non-coding RNAS
All of the following variations mentioned are in comparison to isoform 1 of the human FAM120AOS gene. Isoform 3 is 2199 bp and uses an alternate splice site in the first exon.[2][12] The transcript variants (e.g. isoforms) 6-12 are all candidates for nonsense-mediated mRNA decay (NMD).[2]
Isoform 4 of the gene is 2320 bp and uses an alternate splice site in the first exon and contains an alternate internal exon.[2][13] Isoform 5 is 6043 bp and contains an alternate internal exon. Isoform 6 is 5272 bp and contains an alternate first exon and an alternate internal exon.[2][14] Isoform 7 is 5095 bp and contains an alternate first exon.[2][15] Isoform 8 is 5129 bp and contains an alternate first exon and alternate internal exon.[2][16] Isoform 9 is 5151 bp and contains an alternate first exon.[2][17] Isoform 10 is 5354 bp and contains an alternate first exon.[2][18] Isoform 11 is 5475 bp and contains an alternate first exon and an alternate internal exon.[2][19] Lastly, isoform 12 5216 is bp and contains an alternate first exon and an alternate internal exon.[2]
Proteins
Isoforms
There are two different isoforms of the human FAM120AOS gene that encode a protein, isoforms 1 and 2.[2] The uncharacterized protein FAM120AOS isoform 1 is 256 amino acids long[20] and the uncharacterized protein FAM120AOS isoform 2 is 74 amino acids long.[21] Uncharacterized protein FAM120AOS isoform 1 is the longer and more abundant isoform found in humans, and contains protein domain Q5T035.[2][8] The isoform also has a protein interactant, Q5T035-F120S_HUMAN, and CRISPR reagents and clone products of the protein available.[8]
Molecular weight
Uncharacterized protein FAM120AOS isoform 1 (protein isoform 1) in humans has a calculated molecular weight of 27.8 kDa.[2] A theoretical value of 11.93 for the isoelectric point of the protein was determined through the use of ExPASy.[22] The basic isoelectric point indicates that protein isoform 1 is primarily basic. Table 1 shows the isoelectric points and molecular weights for all the different orthologs of the human FAM120AOS protein 1 across Primates and Artiodactyla.[23] The isoelectric point of the protein remains within a pH of 10.05-11.93 across all orthologs, indicating that the protein is primarily basic. However, the molecular weight of the FAM120AOS protein seems to vary greatly between orthologs, ranging from values of 8.1 kDa to 17.9 kDa, with a maximum value of 29.8 kDa. Many of the sequences with a lower molecular weight were found to be composed of fewer amino acids than the sequences with larger molecular weights. These length differences could also be attributed to possible different isoforms of the FAM120AOS protein being analyzed.
| Organism | Taxonomic Group | Isoelectric Point | Molecular Weight (in kDa) |
| Homo sapiens | Primates | 11.93 | 27.9 |
| Pan troglodytes | Primates | 11.92 | 27.7 |
| Pongo abelii | Primates | 11.69 | 27.8 |
| Nomascus leucogenys | Primates | 10.32 | 7.9 |
| Hylobates moloch | Primates | 10.06 | 8.1 |
| Trachypithecus francoisi | Primates | 11.35 | 8.1 |
| Rhinopithecus roxellana | Primates | 11.57 | 8.3 |
| Macaca nemestrina | Primates | 11.35 | 8.1 |
| Papio anubis | Primates | 10.98 | 8.2 |
| Carlito syrichta | Primates | 11.36 | 25.8 |
| Microcebus murinus | Primates | 11.52 | 29.8 |
| Muntiacus muntjak | Artiodactyla | 11.21 | 17.9 |
Amino acid composition

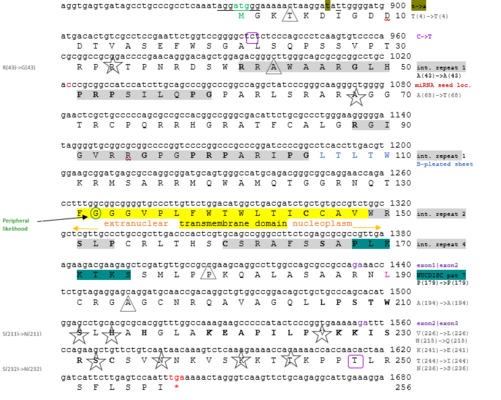
Protein isoform 1 contains two different internal repeats in its amino acid composition, determined through analysis of the protein sequence using Dotlet JS.[24] The first internal repeat occurs at amino acid positions 41-59 and 88-105.[24] The second internal repeat occurs at amino acid positions 145-153 and 160-168.[24] There is an upstream in-frame stop codon (taa) present at amino acid positions 806-808.[2] There is an alternate polyadenylation site present at amino acid positions 2726-2731.[2] The polyadenylation signal used is present present from amino acid positions 5889-5893.[2] The amino acid positions from L206-S211, H213, H215, K219-P225, and K227-C233 were found to be conserved across all of the strict orthologs of the human uncharacterized protein FAM120AOS isoform 1.[25] The amino acid G95 was found to be conserved across all Primates and Artiodactyla for which sequences were identified.[25] The human FAM120AOS protein 1 was found to arginine-rich, and glutamic acid and tyrosine-poor.[26]

Domains and motifs
The uncharacterized protein FAM120AOS isoform 1 in humans contains the protein domain Q5T035.[8]
Two notable motifs found using a eukaryotic linear motif analysis for the human FAM120AOS protein 1 are TRG_RT_diArg_1 and TRG_NLS_MonoExtN_4.[28] The TRG_RT_diArg_1 motif is a di Arginine retention/retrieving signal that is present on membrane proteins, where it serves for ER localization.[28] The TRG_NLS_MonoExtN_4 is a NLS classical nuclear localization signal, which is possessed by many nuclear proteins, indicating that the human FAM120AOS protein 1 is a nuclear protein.
Secondary structure
The secondary structure of the human FAM120AOS protein 1 was predicted by the I-TASSER server and shows 11 alpha helices as follows, in order of position: SER15-TRP18, PRO25-SER27, THR34-TRP40, ALA85-ARG88, LYS111-ALA121, CYS145-ARG155, HIS158-ALA163, LEU169-LYS171, PRO179-ARG198, PRO225-CYS233, and PRO246-PHE252.[29]
Tertiary and quaternary structure

The tertiary structure of the human FAM120AOS protein 1 was predicted by the I-TASSER server with a C-score of -4.00.[29] It appears that the outermost parts of the protein are more solvent accessible, while the inner areas are less solvent accessible.[29] The protein appears to be primarily blue, again indicating that it is a basic structure.[29] The protein also indicated the presence of a peripheral likelihood of 1.48 at amino acid position 132.[30] The NUCDISC results indicated the presence of pat 7 PLKKTKS (4) starting at amino acid position 168.[30]
Gene regulation
Promoter
There are four different promoters for the human FAM120AOS protein 1, which are depicted in the table below.[31] The promoter used for further analysis below (GXP_1829163) is 1665 base pairs long from coordinates 93450944-93452608, with five coding transcripts.[31]
| Promoter | Size (in base pairs) | Coordinates | Strand | Coding Transcripts |
| GXP_9004065 | 1040 | 93437082-93438121 | - | None (non-coding only) |
| GXP_228179 | 1040 | 93446357-93447396 | - | None (non-coding only) |
| GXP_1829163 | 1665 | 93450944-93452608 | - | 5 |
| GXP_2255852 | 1487 | 93453115-93454601 | - | 2 |
Transcription factor binding sites
The transcription factors described below were identified on the Human FAM120A protein 1 promoter.[32]
| Code Name | Full Name | Binding | Matrix Score | Start site | End site |
| AP2F | Activator protein 2 | agcGCCAgacggcac | 0.862 | 336 | 350 |
| STEM | Motif composed of binding sites for pluripotency or stem cell factors | cccgtctGCATggcccact | 0.912 | 255 | 273 |
| ZF20 | C2H2 Zinc finger transcription factors 20 | tgcggttACCA | 0.791 | 447 | 457 |
| E2FF | E2F-myc activator/cell cycle regulator | tggacacggGATAatgg | 0.754 | 29 | 45 |
| ZF5F | ZF5 POZ domain zinc finger | ccctgaGCGCcccaggc | 0.957 | 28 | 44 |
| P53F | P53 tumor suppressor | tgcggttaccaaaggCAAGtcagtg | 0.954 | 312 | 336 |
| RXRF | RXR heterodimer binding sites | ttattgacctagGGTCatattatag | 0.857 | 156 | 180 |
| EBOX | E-box binding factors | attatccCGTGtccaga | 0.901 | 466 | 482 |
| ZF02 | C2H2 Zinc finger transcription factors 2 | caaaagcaCCCCcctacacccgc | 0.933 | 91 | 113 |
| AP1R | MAF and AP1 related factors | ttggttGCTGagaaatttctagtag | 0.842 | 356 | 380 |
| PLAG | pleomorphic adenoma gene | taggGGGGtgcttttgctttcct | 0.871 | 114 | 136 |
| KLFS | Krueppel like transcription factors | agagcttAAAGgattcttc | 0.976 | 118 | 136 |
| ETSF | Human and murine ETS1 factors | ttcagtgaGGAAagcaaaagc | 0.933 | 196 | 216 |
Expression pattern
An immunohistochemical staining of the FAM120AOS protein in the human prostate using a FAM120AOS polyclonal antibody indicates the presence of FAM120AOS in the nucleus of glandular cells.[33]
In Homo sapiens (humans), the gene exhibits high levels of expression (in RPKM) in the colon, fat, placenta, prostate, and thyroid, as determined through quantitative transcriptomic analysis (RNA-Seq) with the following respective values: 12.598, 11.727, 10.978, 11.277, and 13.511.[4] During human fetal development, the gene exhibits the highest levels of expression in the intestine at 20 weeks and the lungs at 17 weeks, as determined through the use of circular RNA with the following respective mean RPKM values: 5.066 and 4.365.[5] The sequencing of RNA from 20 human tissues showed the highest levels of FAM120AOS expression in the placenta, prostate, and thyroid, with respective mean RPKM values of 7.057, 3.978, and 4.396.[7] Transcription profiling through high throughput sequencing of both individual and mixtures of 16 human tissues RNA also found high levels of FAM120AOS gene expression in the thyroid, with a mean RPKM of 9.518.[6]



Transcript level regulation

There are 4 large stem loops present in the 5' UTR of the human FAM120AOS protein 1.[38] There are 8 miRNA binding sites identified for the human FAM120AOS protein 1.[39]
| miRNA Name | miRNA sequence | Target Score | Seed Location |
| hsa-miR-4286 | ACCCCACUCCUGGUACC | 94 | 475 |
| hsa-miR-3059-5p | UUUCCUCUCUGCCCCAUAGGGUGU | 88 | 199, 396 |
| hsa-miR-3152 | UGUGUUAGAAUAGGGGCAAUAA | 87 | 173,735 |
| hsa-miR-4499 | AAGACUGAGAGGAGGGA | 83 | 730 |
| hsa-miR-129-2-3p | AAGCCCUUACCCCAAAAAGCAU | 83 | 1022 |
| hsa-miR-129-1-3p | AAGCCCUUACCCCAAAAAGUAU | 83 | 1022 |
| hsa-miR-6881-3p | AUCCUCUUUCGUCCUUCCCACU | 82 | 199, 395 |
| hsa-miR-10400-3p | CUGGGCUCCCGGACGAGGCGGG | 81 | 337 |
Protein level regulation
The K-NN prediction results for the human FAM120AOS protein 1 predicted it to be present in the nucleus of cells.[30] There is a possible transmembrane domain for the protein, present from amino acid position 131-148.[40]

Homology/evolution
There were no paralogs identified for human FAM120AOS.[2][8] The most distant homolog for human FAM120AOS detectable is the Microcebus murinus, with a 61.17% sequence identity to the human protein.[42] There was a total of 11 orthologs identified for human FAM120AOS protein 1.[43] No proteins with homologous domains to the human FAM120AOS sequence were identified.[44] FAM120AOS seems to be evolving at a moderate rate, in between that of cytochrome c and fibrinogen alpha.[2]
| Genus and species | Common Name | Taxonomic group | Date of divergence (in MYA) | Accession number | Sequence length (in aa) | Sequence Identity to human protein | Sequence similarity to human protein |
| Homo sapiens | Human | Primates | 0 | NP_942138.2 | 256 | 100.00% | 100% |
| Pan troglodytes | Chimpanzee | Primates | 6.4 | PNI17265.1 | 255 | 98.44% | 100% |
| Pongo abelii | Sumtran orangutan | Primates | 15.2 | PNJ71424.1 | 253 | 95.70% | 100% |
| Nomascus leucogenys | Northern white-cheeked gibbon | Primates | 19.8 | XP_030657822.1 | 73 | 94.12% | 26% |
| Hylobates moloch | Silvery gibbon | Primates | 19.8 | XP_032020454.1 | 86 | 92.65% | 26% |
| Trachypithecus francoisi | Francois' leaf monkey | Primates | 28.81 | XP_033092605.1 | 74 | 92.75% | 26% |
| Rhinopithecus roxellana | Golden snub-nosed monkey | Primates | 28.81 | XP_030775307.1 | 74 | 92.75% | 26% |
| Macaca nemestrina | Southern pit-tailed macaque | Primates | 28.81 | XP_024642522.1 | 74 | 92.75% | 26% |
| Papio anubis | Olive baboon | Primates | 28.81 | XP_003912044.1 | 74 | 92.30% | 26% |
| Carlito syrichta | Philippine tarsier | Primates | 69 | XP_021572479.1 | 236 | 66.82% | 78% |
| Microcebus murinus | Mouse lemur | Primates | 74.1 | XP_020144792 | 274 | 61.17% | 76% |
| Muntiacus muntjak | Indian muntjac | Artiodactyla | 94 | KAB0347543.1 | 161 | 97.18% | 27% |


Function/biochemistry
The function and biochemistry of the human FAM120AOS protein are currently unknown.[2][8] The single nucleotide polymorphisms (SNPs) did not show any mutations in conserved amino acids, so it is lis likely that two copies of the FAM120AOS gene are necessary for proper function.
Interacting proteins
The FAM120AOS protein is physically associated with the following proteins: MDFI, ELAV1, TRIM25, and APEX1.[46][47][48][49][8]
Clinical significance
A missense mutation in the FAM120AOS protein from amino acid threonine at position 248 to isoleucine (T248I) has been linked in one whole-of-exome sequencing study to: coarse facial features, scoliosis, pectus excavatum, skin laxity, hypotonia, GERD, hyperreactive airway disease,[lower-alpha 1] and undescended testicles.[50]
Notes
- ↑ Elsewhere, Alazami et al. narrow the description of the reported "chronic lung disease" of this participant to "hyperactive airways" instead.[50] This alternative designation appears in the Supplemental Information of the article.[52] In the main body of the article (at Table 2, pp. 156–157), the phenotype of the individual possessing the candidate gene mutation includes "chronic lung disease". However, chronic lung—or chronic respiratory diseases—are specified by various authorities as particular diseases or conditions, and do not include "hyperactive airways". According to the Open Targets Platform,[53] the thoracic societies of the United States (ATS) and Britain (BTS) list chronic lung disease as: fibrosis, bronchiectasis, bullae, emphysema, and nodular or lymphomatous abnormalities. The Australian Institute of Health and Welfare gives asthma, COPD, allergic rhinitis, bronchiectasis, chronic sinusitis, cystic fibrosis, occupational lung diseases, and pulmonary fibrosis as chronic respiratory conditions.[54]
References
- ↑ 1.0 1.1 1.2 1.3 "AceView: Gene:FAM120AOS, a comprehensive annotation of human, mouse and worm genes with mRNAs or ESTsAceView.". https://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/av.cgi?db=human&term=FAM120AOS&submit=Go.
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 2.15 2.16 2.17 2.18 2.19 2.20 2.21 2.22 2.23 2.24 2.25 2.26 "FAM120AOS family with sequence similarity 120A opposite strand [Homo sapiens (human) - Gene - NCBI"]. https://www.ncbi.nlm.nih.gov/gene/158293#gene-expression.
- ↑ "GDS3834 / 7875". https://www.ncbi.nlm.nih.gov/geo/tools/profileGraph.cgi?ID=GDS3834:7875.
- ↑ 4.0 4.1 "Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics". Molecular & Cellular Proteomics 13 (2): 397–406. February 2014. doi:10.1074/mcp.M113.035600. PMID 24309898.
- ↑ 5.0 5.1 "Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development". Genome Biology 16: 126. June 2015. doi:10.1186/s13059-015-0690-5. PMID 26076956.
- ↑ 6.0 6.1 "Illumina bodyMap2 transcriptome (ID 204271) - BioProject - NCBI". https://www.ncbi.nlm.nih.gov/bioproject/PRJEB2445/.
- ↑ 7.0 7.1 "Genome-wide identification of zero nucleotide recursive splicing in Drosophila". Nature 521 (7552): 376–379. May 2015. doi:10.1038/nature14475. PMID 25970244. Bibcode: 2015Natur.521..376D.
- ↑ 8.0 8.1 8.2 8.3 8.4 8.5 8.6 8.7 "FAM120AOS Gene - F120S Protein - F120S Antibody". https://www.genecards.org/cgi-bin/carddisp.pl?gene=FAM120AOS.
- ↑ NCBI (2020-10-12) (in en-US). Homo sapiens family with sequence similarity 120A opposite strand (FAM120AOS), transcript variant 1, mRNA - NCBI Reference Sequence: NM_198841.4. http://www.ncbi.nlm.nih.gov/nuccore/NM_198841.4.
- ↑ (in en-US) Homo sapiens family with sequence similarity 120A opposite strand (FAM120AOS), transcript variant 1, mRNA. 2020-10-12. http://www.ncbi.nlm.nih.gov/nuccore/NM_198841.4.
- ↑ (in en-US) Homo sapiens family with sequence similarity 120A opposite strand (FAM120AOS), transcript variant 2, mRNA. 2020-12-12. http://www.ncbi.nlm.nih.gov/nuccore/NM_001322224.3.
- ↑ (in en-US) Homo sapiens family with sequence similarity 120A opposite strand (FAM120AOS), transcript variant 3, non-coding RNA. 2020-07-23. http://www.ncbi.nlm.nih.gov/nuccore/NR_136229.2.
- ↑ (in en-US) Homo sapiens family with sequence similarity 120A opposite strand (FAM120AOS), transcript variant 4, non-coding RNA. 2020-07-23. http://www.ncbi.nlm.nih.gov/nuccore/NR_136230.2.
- ↑ (in en-US) Homo sapiens family with sequence similarity 120A opposite strand (FAM120AOS), transcript variant 6, non-coding RNA. 2020-07-25. http://www.ncbi.nlm.nih.gov/nuccore/NR_136232.3.
- ↑ (in en-US) Homo sapiens family with sequence similarity 120A opposite strand (FAM120AOS), transcript variant 7, non-coding RNA. 2020-07-25. http://www.ncbi.nlm.nih.gov/nuccore/NR_136233.3.
- ↑ (in en-US) Homo sapiens family with sequence similarity 120A opposite strand (FAM120AOS), transcript variant 8, non-coding RNA. 2020-07-25. http://www.ncbi.nlm.nih.gov/nuccore/NR_136234.3.
- ↑ (in en-US) Homo sapiens family with sequence similarity 120A opposite strand (FAM120AOS), transcript variant 9, non-coding RNA. 2020-07-25. http://www.ncbi.nlm.nih.gov/nuccore/NR_136235.3.
- ↑ (in en-US) Homo sapiens family with sequence similarity 120A opposite strand (FAM120AOS), transcript variant 10, non-coding RNA. 2020-07-25. http://www.ncbi.nlm.nih.gov/nuccore/NR_136236.3.
- ↑ (in en-US) Homo sapiens family with sequence similarity 120A opposite strand (FAM120AOS), transcript variant 11, non-coding RNA. 2020-07-25. http://www.ncbi.nlm.nih.gov/nuccore/NR_136237.3.
- ↑ "Uncharacterized protein FAM120AOS isoform 1 [Homo sapiens - Protein - NCBI"]. https://www.ncbi.nlm.nih.gov/protein/NP_942138.2.
- ↑ "Uncharacterized protein FAM120AOS isoform 2 [Homo sapiens - Protein - NCBI"]. https://www.ncbi.nlm.nih.gov/protein/NP_001309153.1.
- ↑ "ExPASy: The proteomics server for in-depth protein knowledge and analysis". Nucleic Acids Research 31 (13): 3784–8. July 2003. doi:10.1093/nar/gkg563. PMID 12824418.
- ↑ "Compute pI/MW - SIB Swiss Institute of Bioinformatics". https://www.expasy.org/resources/compute-pi-mw.
- ↑ 24.0 24.1 24.2 "Dotlet JS". https://dotlet.vital-it.ch/.
- ↑ 25.0 25.1 "Clustal Omega < Multiple Sequence Alignment < EMBL-EBI". https://www.ebi.ac.uk/Tools/msa/clustalo/.
- ↑ "EBI Tools". https://www.ebi.ac.uk/Tools/services/web/toolresult.ebi?jobId=saps-I20201206-180725-0931-48392118-p2m.
- ↑ "Clustal Omega < Multiple Sequence Alignment < EMBL-EBI". https://www.ebi.ac.uk/Tools/msa/clustalo/.
- ↑ 28.0 28.1 "ELM". http://elm.eu.org/cgimodel.py?fun=smartResult&userId=QiKRZ3LXL4&EXPECT_CUTOFF=100&r=1&bg=on.
- ↑ 29.0 29.1 29.2 29.3 "I-TASSER server for protein structure and function prediction". https://zhanglab.ccmb.med.umich.edu/I-TASSER/.
- ↑ 30.0 30.1 30.2 "PSORT II Prediction". https://psort.hgc.jp/form2.html.
- ↑ 31.0 31.1 "Gene2Promoter". https://www.genomatix.de/online_help/help_eldorado/Gene2Promoter_Intro.html.
- ↑ "MatInspector: Search for transcription factor binding sites". https://www.genomatix.de/online_help/help_matinspector/matinspector_help.html.
- ↑ "FAM120AOS Antibodies". https://www.thermofisher.com/antibody/primary/target/fam120aos.
- ↑ "GEO Profiles - 104737339 - NCBI". https://www.ncbi.nlm.nih.gov/geoprofiles/104737339.
- ↑ "GEO Profiles - 100460041 - NCBI". https://www.ncbi.nlm.nih.gov/geoprofiles/100460041.
- ↑ "104737339 - GEO Profiles - NCBI". https://www.ncbi.nlm.nih.gov/geoprofiles/104737339.
- ↑ "104737339 - GEO Profiles - NCBI". https://www.ncbi.nlm.nih.gov/geoprofiles/104737339.
- ↑ "RNAfold web server". http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi.
- ↑ 39.0 39.1 "miRDB - Custom Prediction". http://mirdb.org/custom.html.
- ↑ "EBI Tools: Job not available". https://www.ebi.ac.uk/Tools/services/web/toolresult.ebi?jobId=saps-I20201206-180725-0931-48392118-p2m.
- ↑ "104737339 - GEO Profiles - NCBI". https://www.ncbi.nlm.nih.gov/geoprofiles/104737339.
- ↑ "LOW QUALITY PROTEIN: uncharacterized protein FAM120AOS [Microcebus mur - Protein - NCBI". https://www.ncbi.nlm.nih.gov/protein/XP_020144792.
- ↑ "BLAST: Basic Local Alignment Search Tool". https://blast.ncbi.nlm.nih.gov/Blast.cgi.
- ↑ "Human BLAT Search". https://genome.ucsc.edu/cgi-bin/hgBlat.
- ↑ "TimeTree:The Timescale of Life". http://timetree.org/.
- ↑ "MDFI - MyoD family inhibitor, isoform CRA_a - Homo sapiens (Human) - MDFI gene & protein". https://www.uniprot.org/uniprot/A0A024RCY2.
- ↑ "ELAVL1 - ELAV-like protein 1 - Homo sapiens (Human) - ELAVL1 gene & protein" (in en). https://www.uniprot.org/uniprot/Q15717.
- ↑ "TRIM25 Gene - TRI25 Protein -TRI25 Antibody". https://www.genecards.org/cgi-bin/carddisp.pl?gene=TRIM25.
- ↑ "FAM120AOS (RP11-165J3.1) Result Summary - BioGRID". https://thebiogrid.org/127664/summary/homo-sapiens/fam120aos.html.
- ↑ 50.0 50.1 "Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families" (in English). Cell Reports 10 (2): 148–61. January 2015. doi:10.1016/j.celrep.2014.12.015. PMID 25558065.
- ↑ "SNP linked to Gene (geneID:158293) Via Contig Annotation". https://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?chooseRs=coding&locusId=158293&mrna=NM_198841.3&ctg=NT_008470.20&prot=NP_942138.2&orien=reverse&refresh=refresh.
- ↑ "Supplemental Information: Document S1 - Figures S1 and S2 and Table S2 of 'Accelerating novel candidate gene discovery in neurogenetic disorders'" (in English). Cell Reports 10 (2): 148–161. January 2015. doi:10.1016/j.celrep.2014.12.015. Unpaginated: appears under heading "Family: 12DG0321; Gene: FAM120AOS". PMID 25558065. https://www.cell.com/cms/10.1016/j.celrep.2014.12.015/attachment/40067b49-9743-4814-bec3-3bc3bd23b8c3/mmc1.pdf.
- ↑ "Evidence for FAM120AOS in chronic lung disease" (in en). https://platform.opentargets.org/evidence/ENSG00000188938/EFO_0009910.
- ↑ "About Chronic respiratory conditions" (in en-AU). Government of Australia. 28 July 2020. https://www.aihw.gov.au/reports-data/health-conditions-disability-deaths/chronic-respiratory-conditions/about.
