Biology:FAM163A
 Generic protein structure example |
FAM163A, also known as cebelin and neuroblastoma-derived secretory protein (NDSP) is a protein that in humans is encoded by the FAM163A gene.[1] This protein has been implicated in promoting proliferation and anchorage-independent growth of neuroblastoma cancer cells.[2][3] In addition, this protein has been found to be up-regulated in the lung tissue of chronic smokers.[4] FAM163A is found on human chromosome 1q25.2; its protein product is 167 amino acids long. FAM163A contains a very highly conserved signal peptide sequence, coded for by the first ~37 amino acids in its sequence; albeit only conserved in eukaryotes, the most distant of which being the Japanese Rice Fish.
Gene
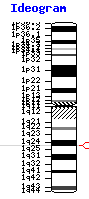
FAM163A is approximately 2,927 base pairs long, containing five exons. While no domains of unknown function have been documented, the coding region of the gene is very short (~500 base pairs), with an exceptionally long and as-of-yet uncharacterized 3' untranslated region (UTR). FAM163A is located on the positive strand of chromosome 1, in loci126860, near three other genes: TOR1AIP1, TOR1AIP2, and TDRD5.[5]
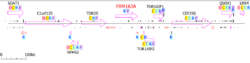
mRNA
mRNA levels were tested in 45 neuroblastoma tumor samples; in 43 of these samples, elevated levels of NDSP were found, as well as in five bone marrow samples. NDSP is associated with increased risk for development of cancer metastasis in bone marrow as well as neural tissue.[2] RNA inhibition techniques applied against NDSP decreased cellular proliferation and cancer cell colony formation. Further, this protein has been determined to act as a growth factor through an ERK-mediated pathway.[3]
Splice variants
Several programs can be used to generate possible splice variants of the Fam163A mRNA. The Ensembl database yields one possible splice variant, which coded for the FAM163A protein.[7] NCBI's Aceview yields 23 possible splice variants, but no experimental evidence is associated with these.[8]
Protein
The human protein has a molecular weight of 17.6 kilodaltons (kDa), and an isoelectric point of 5.56.[9] When compared across orthologs, these values are well conserved. Lastly the ExPASy program PSORTII predicts a 39.1% chance of the protein's localization in the nucleus; this being the highest probability for any location.[10]
| Localization Area | Chances of Localization (%) |
|---|---|
| Nucleus | 39.1% |
| Cytoplasm | 21.7% |
| Extracellular Matrix | 17.4% |
| Mitochondria | 17.4% |
| Cytoskeleton | 4.3% |
Homology
The following data was generated using the NCBI BLAST program.[11] An interesting motif in all of these sequences is the exceptional conservation of the signal peptide sequence; Vasudevan, et al.'s studies included bioinformatic analysis that compared a paralogous protein (FAM163B) in humans and the FAM163A ortholog in mice.[2] Their results aligned with the analysis of the orthologs presented below; while many, many more orthologs exist for FAM163A in species not listed, the Japanese Rice Fish is the last orthologous species that shares the signal peptide sequence, with the next closest result having a percent identity of less than 30% and no putative domains of conservation.
| Genus and species | Common name | Evolutionary time to human divergence (MYA) | Accession # | Protein sequence length | Sequence identity to human protein (%) | Sequence similarity to human protein (%) |
|---|---|---|---|---|---|---|
| Homo sapiens | Human | - | NP_775780.1 | 167aa | - | - |
| Homo sapiens | Human (FAM163B - Paralog) | - | NP_001073984 | 166aa | 42% | 52% |
| Gorilla gorilla gorilla | Gorilla | 8.8 | XP_004028035 | 167aa | 99% | 98% |
| Felis catus | Cat | 94.2 | XP_003999284 | 166aa | 92% | 92% |
| Pteropus alecto | Black Flying Fox | 94.2 | XP_006907838 | 167aa | 89% | 90% |
| Odobenus rosmarus divergens | Pacific Walrus | 94.2 | XP_004398165 | 166aa | 88% | 89% |
| Dasypus novemcinctus | 9-Banded Armadillo | 104.2 | XP_004461936 | 165aa | 87% | 88% |
| Ochotona princeps | American Pika | 92.3 | XP_004598689 | 165aa | 86% | 89% |
| Mus musculus | Mouse | 92.3 | Q8CAA5 | 168aa | 85% | 87% |
| Alligator mississippiensis | American Alligator | 296 | XP_006276882 | 161aa | 66% | 74% |
| Pelodiscus sinensis | Chinese Soft-Shelled Turtle | 296 | XP_004461936 | 164aa | 64% | 73% |
| Gallus gallus | Chicken | 296 | XP_001234382 | 159aa | 61% | 67% |
| Ophiophagus hannah | King Cobra | 296 | ETE64717 | 166aa | 53% | 65% |
| Danio rerio | Zebrafish | 400.1 | XP_002660900 | 150aa | 50% | 63% |
| Xiphophorus maculatus | Southern Platyfish | 400.1 | XP_005800930 | 163aa | 48% | 60% |
| Oryzias latipes | Japanese Rice Fish | 400.1 | XP_004067975 | 163aa | 46% | 60% |
Paralogs
FAM163A has only one paralog: FAM163B, located on chromosome 9q34.2. Comparison between the two proteins reveals that the signal peptide sequence is identical; using the CLUSTALW program through SDSC's Biology Workbench, it was possible to visualize the sequences' identity.[12]
Tissue distribution
FAM163A is ubiquitously expressed at very low levels in most tissues of the body; expression is higher in juveniles, and as previously seen, in chronic smokers' lungs and neuroblastoma cells.[13]
References
- ↑ "FAM163a Gene". GeneCards. Weizmann Institute of Science. https://www.genecards.org/cgi-bin/carddisp.pl?gene=FAM163A&search=FAM163A.
- ↑ 2.0 2.1 2.2 "Neuroblastoma-derived secretory protein is a novel secreted factor overexpressed in neuroblastoma". Mol. Cancer Ther. 8 (8): 2478–89. 2009. doi:10.1158/1535-7163.MCT-08-1132. PMID 19671756.
- ↑ 3.0 3.1 "Neuroblastoma-derived secretory protein messenger RNA levels correlate with high-risk neuroblastoma". J. Pediatr. Surg. 42 (1): 148–52. 2007. doi:10.1016/j.jpedsurg.2006.09.064. PMID 17208556.
- ↑ Tobacco and Genetics Consortium (2010). "Genome-wide meta-analyses identify multiple loci associated with smoking behavior". Nat. Genet. 42 (5): 441–7. doi:10.1038/ng.571. PMID 20418890.
- ↑ "NCBI Gene". https://www.ncbi.nlm.nih.gov/gene?LinkName=nuccore_gene&from_uid=34222229.
- ↑ "NCBI AceView". NCBI. https://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/av.cgi?db=human&term=FAM163A&submit=Go.
- ↑ "Gene: FAM163A". Ensembl Genome Browser. Wellcome Trust Genome Campus. http://useast.ensembl.org/Homo_sapiens/Gene/Splice?db=core;g=ENSG00000143340;r=1:179712298-179785333;t=ENST00000341785.
- ↑ "AceView: FAM163A". NCBI's AceView. https://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/av.cgi?db=human&term=FAM163A&submit=Go.
- ↑ "ExPASy: pI/Mw". ExPASy. CBS. http://www.cbs.dtu.dk/cgi-bin/webface2.fcgi?jobid=53770B9E0000648E8EE5E200&wait=20.
- ↑ "ExPASy: PSORTII". ExPASy. CBS. http://www.cbs.dtu.dk/cgi-bin/webface2.fcgi?jobid=53770B9E0000648E8EE5E200&wait=20.
- ↑ "NCBI BLAST". NCBI. http://blast.ncbi.nlm.nih.gov/Blast.cgi.
- ↑ "CLUSTALW". SDSC Biology Workbench. University of California, San Diego. http://workbench.sdsc.edu/.
- ↑ "NCBI GEO: mining tens of millions of expression profiles--database and tools update". Nucleic Acids Res. 35 (Database issue): D760–5. January 2007. doi:10.1093/nar/gkl887. PMID 17099226.
Further reading
- "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides.". Gene 138 (1–2): 171–4. 1994. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- "DNA cloning using in vitro site-specific recombination". Genome Res. 10 (11): 1788–95. 2001. doi:10.1101/gr.143000. PMID 11076863.
- "A protein interaction framework for human mRNA degradation". Genome Res. 14 (7): 1315–23. 2004. doi:10.1101/gr.2122004. PMID 15231747.
- "The LIFEdb database in 2006". Nucleic Acids Res. 34 (Database issue): D415–8. 2006. doi:10.1093/nar/gkj139. PMID 16381901.
 |
