Biology:Ferulic acid decarboxylase
| AnFDC1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Dimer of Fdc1 from Aspergillus niger. PDB file : 4ZA4 | |||||||||
| Identifiers | |||||||||
| EC number | 4.1.1.102 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
Ferulic acid decarboxylases (Fdc) are decarboxylase enzymes capable of the reversible decarboxylation of aromatic carboxylic acids such as ferulic acid and cinnamic acid.[1] Fdc's are fungal homologues of the E.coli UbiD enzyme [2] which is involved in ubiquinone biosynthesis.[3] This places Fdc within the wider UbiD enzyme family, representing a distinct clade within the family [2] Presence of fdc1 and the associated pad1 genes (Pad1 homologous to UbiX in E.coli) were shown to be required for the decarboxylation of phenylacrylic acids in Saccharomyces cerevisiae.[4]
In 2015 the cofactor prFMN was discovered in the active site of Fdc1 from Aspergillus niger (AnFdc) by crystallography,[5] prior to this genetic studies had led to the assumption that both UbiD and UbiX encoded isofunctional decarboxylases.[6] In actuality UbiX/Pad were found to be flavin preyltransferases[7] supplying the prFMN cofactor to UbiD/Fdc[5] where it is utilised for the reversible decarboxylation of alpha-beta unsaturated carboxylic acid substrates.[2] Since the discovery of prFMN AnFDC has become the most well understood representative of the UbiD enzyme family [8]
AnFDC Mechanism
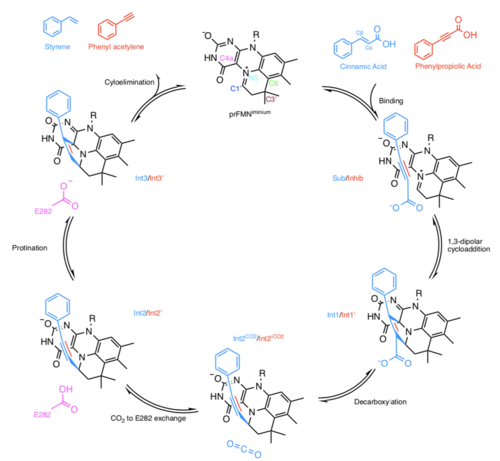
In the same paper in which the structure of prFMN was deduced in the active site of AnFdc1 there was a proposal for the mechanism by which Fdc1 decarboxylates α,β-unsaturated carboxylic acids.[5] Not all UbiD enzymes decarboxylate acrylic acid substrates and other mechanisms may be at play for alternative substrates.[9] In the case of AnFdc1 it was noted that prFMN displays an azomethine ylide characteristic C4a-N5+=C1’(Figure 1). This is a well-known 1,3-dipole in organic chemistry, positioned in the enzyme active site near to the α,β-unsaturated carboxylic acid substrate which contains a 1,3-dipolarophile. Thus, it was proposed that a 1,3-dipolar cycloaddition mechanism was responsible for the enzymatic decarboxylation. This was confirmed in a later paper.[8]
The mechanism proposed in [5] for 1,3-dipolar cycloaddition by Fdc1 is as follows (intermediates represented in Figure 1):
- 1,3-dipolar cycloaddition between prFMNiminium and the α,β-unsaturated substrate leads to a pyrrolidine cycloadduct (Int1)
- This pyrrolidine cycloadduct supports simultaneous decarboxylation and ring opening, resulting in the formation of a distinct prFMN-alkene adduct (Int2)
- A conserved glutamic acid residue (E282) donates a proton to the alkene moiety, resulting a second pyrrolidine cycloadduct (Int3)
- The reaction concludes with cycloelimination of Int3 and the release of the alkene product and CO2
A study went on to present evidence for the 1,3-dipolar cycloaddition,[8] due to suspected turnover of cinnamic acid a crystal structure of AnFdc1 in complex with α-fluorocinnamic acid revealed the substrate Cα and Cβ carbons are located directly above the prFMNiminium C1’ and C4a respectively (shown as Sub in Figure 1 - with cinnamic acid as opposed to α-fluorocinnamic acid). Cinnamic acid was confirmed to bind in a similar manner using inactive AnFdc1 crystals containing FMN. The AnFdc1 E282Q mutant crystallised with cinnamic acid revealed a structure corresponding to the Int2 species, this was taken to mean that progression through the 1,3-dipolarcycloadition cycle was halted as E282 is unable to donate a proton to the alkene moiety.
In order to observe the Int1 and Int3 structures alkyne analogues were used. Like alkenes these compounds can also act as dipolarophiles but cycloaddition would yield a cycloadduct containing a double bond. An inactive AnFdc1 enzyme (with prFMNradical bound) co-crystallised with the phenylpropiolic acid revealed binding in a similar manner to the α-fluorocinnamic acid AnFdc1 and cinnamic acid AnFdc1 with FMN bound (Inhib). An active AnFdc1 enzyme co-crystallised with phenylpropiolic acid revealed clear density for a 3-pyrroline cycloadduct (Int3’) between the alkyne and prFMNiminium. Int3’ represents a structure post decarboxylation, so it was assumed that over the time it took for crystallisation (~24h) the decarboxylation had occurred. Using a rapid soaking procedure, a different cycloadduct was observed that retained the carboxyl moiety (Int1’).
References
- ↑ "FDC1 - Ferulic acid decarboxylase 1 - Aspergillus niger". UniProt. https://www.uniprot.org/uniprot/A0A3F3RNU4.
- ↑ 2.0 2.1 2.2 "The UbiX-UbiD system: The biosynthesis and use of prenylated flavin (prFMN)". Archives of Biochemistry and Biophysics 632: 209–221. October 2017. doi:10.1016/j.abb.2017.07.014. PMID 28754323. https://www.research.manchester.ac.uk/portal/en/publications/the-ubixubid-system(f59b02e9-7372-4bda-a05e-bdda394a560a).html.
- ↑ "Biosynthesis and physiology of coenzyme Q in bacteria". Biochimica et Biophysica Acta (BBA) - Bioenergetics 1837 (7): 1004–1011. July 2014. doi:10.1016/j.bbabio.2014.01.015. PMID 24480387.
- ↑ "PAD1 and FDC1 are essential for the decarboxylation of phenylacrylic acids in Saccharomyces cerevisiae". Journal of Bioscience and Bioengineering 109 (6): 564–569. June 2010. doi:10.1016/j.jbiosc.2009.11.011. PMID 20471595.
- ↑ 5.0 5.1 5.2 5.3 "New cofactor supports α,β-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition". Nature 522 (7557): 497–501. June 2015. doi:10.1038/nature14560. PMID 26083754. Bibcode: 2015Natur.522..497P.
- ↑ "Regulation of the isofunctional genes ubiD and ubiX of the ubiquinone biosynthetic pathway of Escherichia coli". FEMS Microbiology Letters 223 (1): 67–72. June 2003. doi:10.1016/S0378-1097(03)00343-4. PMID 12799002.
- ↑ "UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis". Nature 522 (7557): 502–506. June 2015. doi:10.1038/nature14559. PMID 26083743. Bibcode: 2015Natur.522..502W.
- ↑ 8.0 8.1 8.2 "Enzymatic control of cycloadduct conformation ensures reversible 1,3-dipolar cycloaddition in a prFMN-dependent decarboxylase". Nature Chemistry 11 (11): 1049–1057. November 2019. doi:10.1038/s41557-019-0324-8. PMID 31527849. Bibcode: 2019NatCh..11.1049B.
- ↑ "Flavin metamorphosis: cofactor transformation through prenylation". Current Opinion in Chemical Biology 47: 117–125. December 2018. doi:10.1016/j.cbpa.2018.09.024. PMID 30326424.
 |