Biology:Gal4 transcription factor
This article has multiple issues. Please help improve it or discuss these issues on the talk page. (Learn how and when to remove these template messages)
(Learn how and when to remove this template message) |
| Regulatory protein GAL4 | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Organism | |||||||
| Symbol | GAL4 | ||||||
| Entrez | 855828 | ||||||
| UniProt | P04386 | ||||||
| |||||||
The Gal4 transcription factor is a positive regulator of gene expression of galactose-induced genes.[1] This protein represents a large fungal family of transcription factors, Gal4 family, which includes over 50 members in the yeast Saccharomyces cerevisiae e.g. Oaf1, Pip2, Pdr1, Pdr3, Leu3.[2]
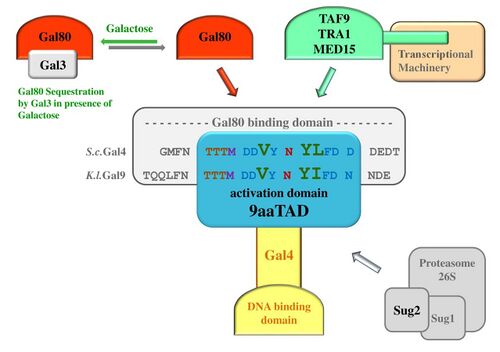
Gal4 recognizes genes with UASG, an upstream activating sequence, and activates them. In yeast cells, the principal targets are GAL1 (galactokinase), GAL10 (UDP-glucose 4-epimerase), and GAL7 (galactose-1-phosphate uridylyltransferase), three enzymes required for galactose metabolism. This binding has also proven useful in constructing the GAL4/UAS system, a technique for controlling expression in insects.[3] In yeast, Gal4 is by default repressed by Gal80, and activated in the presence of galactose as Gal3 binds away Gal80.[4]
Domains
Two executive domains, DNA binding and activation domains, provide key function of the Gal4 protein conforming to most of the transcription factors.
DNA binding
Gal4 N-terminus is a zinc finger and belongs to the Zn(2)-C6 fungal family. It forms a Zn – cysteines thiolate cluster,[5][6] and specifically recognizes UASG in GAL1 promoter. [7][8]
Gal4 transactivation
Localised to the C-terminus, belongs to the nine amino acids transactivation domain family, 9aaTAD, together with Oaf1, Pip2, Pdr1, Pdr3, but also p53, E2A, MLL.[9][10]
Regulation
Galactose induces Gal4 mediated transcription albeit Glucose causes severe repression.[11][12]
As a part of the Gal4 regulation, inhibitory protein Gal80 recognises and binds to the Gal4 region (853-874 aa).[13][14][15]
The inhibitory protein Gal80 is sequestered by regulatory protein Gal3 in Galactose dependent manner. This allows for Gal4 to work when there is galactose.[16][4][17][18]
Mutants
The Gal4 loss-of-function mutant gal4-64 (1-852 aa, deletion of the Gal4 C-terminal 29 aa) lost both interaction with Gal80 and activation function.[19][20][21]
In the Gal4 reverted mutant Gal4C-62 mutant,[22] a sequence (QTAY N AFMN) with the 9aaTAD pattern emerged and restored activation function of the Gal4 protein.
Inactive constructs
The activation domain Gal4 is inhibited by C-terminal domain in some Gal4 constructs.[23][24]
Function
Target
Transcription
The Gal4 activation function is mediated by MED15 (Gal11).[25][26][27][28][29][30][31]
The Gal4 protein interacts also with other mediators of transcription as are Tra1,[32][33][34] TAF9,[35] and SAGA/MED15 complex.[36][37]
Proteosome
A subunit of the 26 S proteasome Sug2 regulatory protein has a molecular and functional interaction with Gal4 function.[38][39] Proteolytic turnover of the Gal4 transcription factor is not required for function in vivo.[40] The native Gal4 monoubiquitination protects from 19S-mediated destabilizing under inducing conditions.[41]
Application
The broad use of the Gal4 is in yeast two-hybrid screening to screen or to assay protein-protein interactions in eukaryotic cells from yeast to human.
In the GAL4/UAS system, the Gal4 protein and Gal4 upstream activating region (UAS) are used to study the gene expression and function in organisms such as the fruit fly.[3]
The Gal4 and inhibitory protein Gal80 have found application in a genetics technique for creating individually labeled homozygous cells called MARCM (Mosaic analysis with a repressible cell marker).
See also
References
- ↑ "Studies on the positive regulatory gene, GAL4, in regulation of galactose catabolic enzymes in Saccharomyces cerevisiae". Molecular & General Genetics 135 (3): 203–12. 1974. doi:10.1007/BF00268616. PMID 4376212.
- ↑ "Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators". Nucleic Acids Research 24 (23): 4599–607. December 1996. doi:10.1093/nar/24.23.4599. PMID 8967907.
- ↑ 3.0 3.1 "GAL4 system in Drosophila: a fly geneticist's Swiss army knife". Genesis 34 (1–2): 1–15. 2002. doi:10.1002/gene.10150. PMID 12324939.
- ↑ 4.0 4.1 "Gene activation by dissociation of an inhibitor from a transcriptional activation domain". Molecular and Cellular Biology 29 (20): 5604–10. October 2009. doi:10.1128/MCB.00632-09. PMID 19651897.
- ↑ "DNA recognition by GAL4: structure of a protein-DNA complex". Nature 356 (6368): 408–14. April 1992. doi:10.1038/356408a0. PMID 1557122. Bibcode: 1992Natur.356..408M.
- ↑ "The DNA binding domain of GAL4 forms a binuclear metal ion complex". Biochemistry 29 (12): 2023–9. March 1990. doi:10.1021/bi00464a019. PMID 2186803.
- ↑ "Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein". Science 231 (4739): 699–704. February 1986. doi:10.1126/science.3080805. PMID 3080805. Bibcode: 1986Sci...231..699K.
- ↑ "Specific DNA binding of GAL4, a positive regulatory protein of yeast". Cell 40 (4): 767–74. April 1985. doi:10.1016/0092-8674(85)90336-8. PMID 3886158.
- ↑ "The DNA binding and activation domains of Gal4p are sufficient for conveying its regulatory signals". Molecular and Cellular Biology 17 (5): 2538–49. May 1997. doi:10.1128/MCB.17.5.2538. PMID 9111323.
- ↑ "GAL4 interacts with TATA-binding protein and coactivators". Molecular and Cellular Biology 15 (5): 2839–48. May 1995. doi:10.1128/MCB.15.5.2839. PMID 7739564.
- ↑ "Studies on the positive regulatory gene, GAL4, in regulation of galactose catabolic enzymes in Saccharomyces cerevisiae". Molecular & General Genetics 135 (3): 203–12. 1974. doi:10.1007/BF00268616. PMID 4376212.
- ↑ "Regulated expression of the GAL4 activator gene in yeast provides a sensitive genetic switch for glucose repression". Proceedings of the National Academy of Sciences of the United States of America 88 (19): 8597–601. October 1991. doi:10.1073/pnas.88.19.8597. PMID 1924319. Bibcode: 1991PNAS...88.8597G.
- ↑ "NADP regulates the yeast GAL induction system". Science 319 (5866): 1090–2. February 2008. doi:10.1126/science.1151903. PMID 18292341. Bibcode: 2008Sci...319.1090K.
- ↑ "The interaction between an acidic transcriptional activator and its inhibitor. The molecular basis of Gal4p recognition by Gal80p". The Journal of Biological Chemistry 283 (44): 30266–72. October 2008. doi:10.1074/jbc.M805200200. PMID 18701455.
- ↑ "Interaction of positive and negative regulatory proteins in the galactose regulon of yeast". Cell 50 (1): 143–6. July 1987. doi:10.1016/0092-8674(87)90671-4. PMID 3297350.
- ↑ "Rapid GAL gene switch of Saccharomyces cerevisiae depends on nuclear Gal3, not nucleocytoplasmic trafficking of Gal3 and Gal80". Genetics 189 (3): 825–36. November 2011. doi:10.1534/genetics.111.131839. PMID 21890741.
- ↑ "Gene activation by interaction of an inhibitor with a cytoplasmic signaling protein". Proceedings of the National Academy of Sciences of the United States of America 99 (13): 8548–53. June 2002. doi:10.1073/pnas.142100099. PMID 12084916. Bibcode: 2002PNAS...99.8548P.
- ↑ "Dilution kinetic studies of yeast populations: in vivo aggregation of galactose utilizing enzymes and positive regulator molecules". Genetics 77 (3): 491–505. July 1974. doi:10.1093/genetics/77.3.491. PMID 4369925.
- ↑ "The genetic control of galactose utilization in Saccharomyces". Journal of Bacteriology 68 (6): 662–70. December 1954. doi:10.1128/jb.68.6.662-670.1954. PMID 13221541.
- ↑ "Enzymatic Expression and Genetic Linkage of Genes Controlling Galactose Utilization in Saccharomyces". Genetics 49 (5): 837–44. May 1964. doi:10.1093/genetics/49.5.837. PMID 14158615.
- ↑ "Function of positive regulatory gene gal4 in the synthesis of galactose pathway enzymes in Saccharomyces cerevisiae: evidence that the GAL81 region codes for part of the gal4 protein". Journal of Bacteriology 141 (2): 508–27. February 1980. doi:10.1128/JB.141.2.508-527.1980. PMID 6988385.
- ↑ "Interaction of positive and negative regulatory proteins in the galactose regulon of yeast". Cell 50 (1): 143–6. July 1987. doi:10.1016/0092-8674(87)90671-4. PMID 3297350.
- ↑ "Deletion analysis of GAL4 defines two transcriptional activating segments". Cell 48 (5): 847–53. March 1987. doi:10.1016/0092-8674(87)90081-X. PMID 3028647.
- ↑ "A sequence-specific transcription activator motif and powerful synthetic variants that bind Mediator using a fuzzy protein interface". Proceedings of the National Academy of Sciences of the United States of America 111 (34): E3506-13. August 2014. doi:10.1073/pnas.1412088111. PMID 25122681. Bibcode: 2014PNAS..111E3506W.
- ↑ "The Saccharomyces cerevisiae SPT13/GAL11 gene has both positive and negative regulatory roles in transcription". Molecular and Cellular Biology 9 (12): 5602–9. December 1989. doi:10.1128/MCB.9.12.5602. PMID 2685570.
- ↑ "Peptides selected to bind the Gal80 repressor are potent transcriptional activation domains in yeast". The Journal of Biological Chemistry 275 (20): 14979–84. May 2000. doi:10.1074/jbc.275.20.14979. PMID 10809742.
- ↑ "Regulation of expression of the galactose gene cluster in Saccharomyces cerevisiae. Isolation and characterization of the regulatory gene GAL4". Molecular & General Genetics 191 (1): 31–8. 1983. doi:10.1007/BF00330886. PMID 6350827.
- ↑ "GAL11 (SPT13), a transcriptional regulator of diverse yeast genes, affects the phosphorylation state of GAL4, a highly specific transcriptional activator". Molecular and Cellular Biology 11 (4): 2311–4. April 1991. doi:10.1128/MCB.11.4.2311. PMID 2005915.
- ↑ "A novel mutation that affects utilization of galactose in Saccharomyces cerevisiae". Current Genetics 2 (2): 115–20. October 1980. doi:10.1007/BF00420623. PMID 24189802.
- ↑ "Yeast GAL11 protein is a distinctive type transcription factor that enhances basal transcription in vitro". Proceedings of the National Academy of Sciences of the United States of America 90 (18): 8382–6. September 1993. doi:10.1073/pnas.90.18.8382. PMID 8378310. Bibcode: 1993PNAS...90.8382S.
- ↑ "GAL11 protein, an auxiliary transcription activator for genes encoding galactose-metabolizing enzymes in Saccharomyces cerevisiae". Molecular and Cellular Biology 12 (10): 4806. October 1992. doi:10.1128/MCB.12.10.4806. PMID 1406662.
- ↑ "Analysis of Gal4-directed transcription activation using Tra1 mutants selectively defective for interaction with Gal4". Proceedings of the National Academy of Sciences of the United States of America 109 (6): 1997–2002. February 2012. doi:10.1073/pnas.1116340109. PMID 22308403. PMC 3277556. Bibcode: 2012PNAS..109.1997L. http://escholarship.umassmed.edu/cgi/viewcontent.cgi?article=1180&context=pgfe_pp.
- ↑ "Tra1 as a screening target for transcriptional activation domain discovery". Bioorganic & Medicinal Chemistry Letters 19 (14): 3733–5. July 2009. doi:10.1016/j.bmcl.2009.05.045. PMID 19497740.
- ↑ "Targets of the Gal4 transcription activator in functional transcription complexes". Molecular and Cellular Biology 25 (20): 9092–102. October 2005. doi:10.1128/MCB.25.20.9092-9102.2005. PMID 16199885.
- ↑ "Use of a genetically introduced cross-linker to identify interaction sites of acidic activators within native transcription factor IID and SAGA". The Journal of Biological Chemistry 278 (9): 6779–86. February 2003. doi:10.1074/jbc.M212514200. PMID 12501245.
- ↑ "The Saccharomyces cerevisiae Srb8-Srb11 complex functions with the SAGA complex during Gal4-activated transcription". Molecular and Cellular Biology 25 (1): 114–23. January 2005. doi:10.1128/MCB.25.1.114-123.2005. PMID 15601835. (http://mcb.asm.org/content/25/1/114/F8.large.jpg)
- ↑ "Functional studies of the yeast med5, med15 and med16 mediator tail subunits". PLOS ONE 8 (8): e73137. 2013. doi:10.1371/journal.pone.0073137. PMID 23991176. Bibcode: 2013PLoSO...873137L.
- ↑ "The Gal4 activation domain binds Sug2 protein, a proteasome component, in vivo and in vitro". The Journal of Biological Chemistry 276 (33): 30956–63. August 2001. doi:10.1074/jbc.M102254200. PMID 11418596.
- ↑ "Evidence that proteolysis of Gal4 cannot explain the transcriptional effects of proteasome ATPase mutations". The Journal of Biological Chemistry 276 (13): 9825–31. March 2001. doi:10.1074/jbc.M010889200. PMID 11152478.
- ↑ "Proteolytic turnover of the Gal4 transcription factor is not required for function in vivo". Nature 442 (7106): 1054–7. August 2006. doi:10.1038/nature05067. PMID 16929306. Bibcode: 2006Natur.442.1054N.
- ↑ "The role of the proteasomal ATPases and activator monoubiquitylation in regulating Gal4 binding to promoters". Genes & Development 21 (1): 112–23. January 2007. doi:10.1101/gad.1493207. PMID 17167105.
Further reading
- Gal4p on WikiGenes
- "Yeast Gal4: a transcriptional paradigm revisited". EMBO Reports 7 (5): 496–9. May 2006. doi:10.1038/sj.embor.7400679. PMID 16670683.
 |