Biology:Gapmer
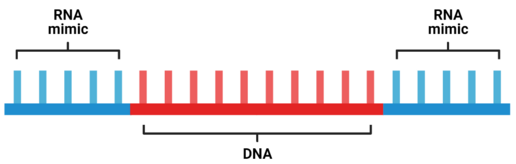
Gapmers are short DNA antisense oligonucleotide structures with RNA-like segments on both sides of the sequence.[1] These linear pieces of genetic information are designed to hybridize to a target piece of RNA and silence the gene through the induction of RNase H cleavage. Binding of the gapmer to the target has a higher affinity due to the modified RNA flanking regions, as well as resistance to degradation by nucleases.[2][3] Gapmers are currently being developed as therapeutics for a variety of cancers, viruses, and other chronic genetic disorders.[4]
Chemical Structure
Gapmers are composed of short DNA strands flanked by strands of RNA mimics. The mimics are typically composed of locked nucleic acids (LNA), 2'-OMe, or 2'-F modified bases.[2] LNA sequences are RNA analogues "locked" into an ideal Watson-Crick base pairing conformation. Gapmers often utilize nucleotides modified with phosphorothioate (PS) groups.[4][3]
Mechanism of Action
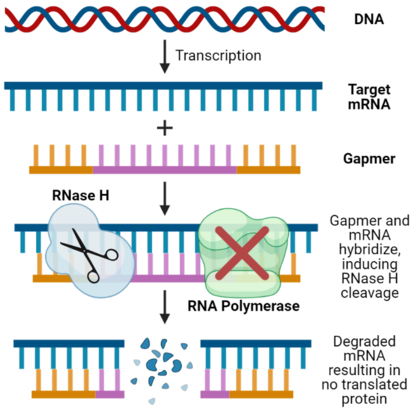
The mechanism of therapeutic gene-silencing action relies on degradation through the action of RNase H.[1][5] Nearly all organisms utilize this family of enzymes to degrade DNA-RNA hybrids as a defense against viral infection.[6] In protein synthesis, DNA is first transcribed into mRNA, and then translated in an amino acid sequence. Gapmers take advantage of this biological pathway by binding to the mRNA target.[7] In humans, the gapmer DNA-mRNA duplex is degraded by RNase H1.[1][5] The degradation of the mRNA prevents protein synthesis[7][8]
Advantages
The gapmer chemical structure is designed to increase resistance to nuclease degradation and enhance stability in vivo.[1][2] LNAs, 2'-OMe, or 2'-F modified bases are chemical analogs of natural RNA nucleic acids. These modifications allow for an increase in nuclease resistance, reduced immunogenicity, and a decrease in toxicity.[1] Gapmers can also have a high binding affinity to the target mRNA.[1] This high binding affinity reduces off-target effects, non-specific binding, and unwanted gene silencing[2]
Therapeutics
Mipomersen (Kynamro)
Kynamro was approved by the FDA in January 2013 for the treatment of homozygous familial hypercholesterolemia (HoFH).[9] The drug, developed by Ionis Pharmaceuticals and marketed by Genzyme Corporation, is administered via subcutaneous injection in the form of a mipomersen sodium solution. The chemical structure is composed of a 20-nucleotide (20-mer) chain with phosphorothioate (PS) backbone modifications and 2'-O-Methoxyethyl (MOE) ribose substitutions.[4] Kynamro targets the mRNA product of the APOB gene, which codes for the Apolipoprotein B-100 protein, a component of low-density lipoprotein (LDL).[2] The binding of mipomersen to the APOB mRNA effectively blocks the translation of ApoB-100, and the gapmer-RNA hybrid is then degraded by the RNase H enzyme. Kynamro is reported to have an elimination half-life of approximately 1–2 months[9]
Inotersen (Tegsedi)
Tegsedi, developed and marketed by Ionis Pharmaceuticals, was approved by the FDA in October 2018 for the treatment of hereditary transthyretin amyloidosis (hATTR).[10] The chemical structure is a 20-mer oligonucleotide with PS backbone modifications and 2'-MOE ribose substitutions.[4] Tegsedi, in the form of an inotersen sodium solution, is administered subcutaneously on a weekly interval. Inotersen binds to the mRNA coding for the transthyretin protein,[11] which blocks translation of the mRNA and recruits RNase enzymes to degrade the gapmer-RNA hybrid. This effectively reduces the level of transthyretin in blood serum, which has been shown to treat polyneuropathy symptoms in patients with hATTR[11]
Safety
Gapmer antisense oligonucleotides (ASOs) have the potential to cause unintended, off-target effects. These off-target effects are produced when the gapmer binds to mRNA with a sufficient degree of complementarity to the target mRNA, blocking or down-regulating the translation of unintended proteins.[12] The functional consequences of gapmer off-target effects can vary widely, depending on the proteins affected and the extent of the down-regulation. Gapmer-based therapeutics also have the potential for side effects. For example, Kynamro has been shown to induce injection site reactions, nausea, headaches, flu-like symptoms, and hepatotoxic reactions.[9] Side effects of Inotersen include thrombocytopenia, glomerulonephritis, injection site reactions, nausea, headache, fatigue, and fever[10]
See also
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Crooke, Stanley T.; Baker, Brenda F.; Crooke, Rosanne M.; Liang, Xue-hai (2021-03-24). "Antisense technology: an overview and prospectus" (in en). Nature Reviews Drug Discovery 20 (6): 427–453. doi:10.1038/s41573-021-00162-z. ISSN 1474-1784. PMID 33762737. https://www.nature.com/articles/s41573-021-00162-z.
- ↑ 2.0 2.1 2.2 2.3 2.4 Dhuri, Karishma; Bechtold, Clara; Quijano, Elias; Pham, Ha; Gupta, Anisha; Vikram, Ajit; Bahal, Raman (2020-06-26). "Antisense Oligonucleotides: An Emerging Area in Drug Discovery and Development". Journal of Clinical Medicine 9 (6): 2004. doi:10.3390/jcm9062004. ISSN 2077-0383. PMID 32604776.
- ↑ 3.0 3.1 QIAGEN. (2017). Antisense LNA GapmeRs Handbook: LNA-optimized oligonucleotides for strand-specific knockdown of mRNA and IncRNA. Germantown, MD: Author
- ↑ 4.0 4.1 4.2 4.3 Roberts, Thomas C.; Langer, Robert; Wood, Matthew J. A. (October 2020). "Advances in oligonucleotide drug delivery" (in en). Nature Reviews Drug Discovery 19 (10): 673–694. doi:10.1038/s41573-020-0075-7. ISSN 1474-1784. PMID 32782413.
- ↑ 5.0 5.1 Kasuya, Takeshi; Hori, Shin-ichiro; Watanabe, Ayahisa; Nakajima, Mado; Gahara, Yoshinari; Rokushima, Masatomo; Yanagimoto, Toru; Kugimiya, Akira (2016-07-27). "Ribonuclease H1-dependent hepatotoxicity caused by locked nucleic acid-modified gapmer antisense oligonucleotides" (in en). Scientific Reports 6 (1): 30377. doi:10.1038/srep30377. ISSN 2045-2322. PMID 27461380. Bibcode: 2016NatSR...630377K.
- ↑ Cerritelli, Susana M.; Crouch, Robert J. (March 2009). "Ribonuclease H: the enzymes in Eukaryotes". The FEBS Journal 276 (6): 1494–1505. doi:10.1111/j.1742-4658.2009.06908.x. ISSN 1742-464X. PMID 19228196.
- ↑ 7.0 7.1 Amodio, Nicola; Stamato, Maria Angelica; Juli, Giada; Morelli, Eugenio; Fulciniti, Mariateresa; Manzoni, Martina; Taiana, Elisa; Agnelli, Luca et al. (September 2018). "Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity" (in en). Leukemia 32 (9): 1948–1957. doi:10.1038/s41375-018-0067-3. ISSN 1476-5551. PMID 29487387.
- ↑ Cheng, Xinwei; Liu, Qibing; Li, Hong; Kang, Chen; Liu, Yang; Guo, Tianqi; Shang, Ke; Yan, Chengyun et al. (2017-02-01). "Lipid Nanoparticles Loaded with an Antisense Oligonucleotide Gapmer Against Bcl-2 for Treatment of Lung Cancer" (in en). Pharmaceutical Research 34 (2): 310–320. doi:10.1007/s11095-016-2063-5. ISSN 1573-904X. PMID 27896589. https://doi.org/10.1007/s11095-016-2063-5.
- ↑ 9.0 9.1 9.2 Mipomersen [package insert]. Cambridge, MA: Genzyme Corporation; 2013.
- ↑ 10.0 10.1 Inotersen [package insert]. Carlsbad, CA: Ionis Pharmaceuticals, Inc.; 2018.
- ↑ 11.0 11.1 "Drug Information Portal - U.S. National Library of Medicine - Quick Access to Quality Drug Information". https://druginfo.nlm.nih.gov/drugportal/name/inotersen.
- ↑ Yoshida, Tokuyuki; Naito, Yuki; Yasuhara, Hidenori; Sasaki, Kiyomi; Kawaji, Hideya; Kawai, Jun; Naito, Mikihiko; Okuda, Haruhiro et al. (2019). "Evaluation of off-target effects of gapmer antisense oligonucleotides using human cells" (in en). Genes to Cells 24 (12): 827–835. doi:10.1111/gtc.12730. ISSN 1365-2443. PMID 31637814.
This article needs additional or more specific categories. (June 2021) |
 |