Biology:Gene-environment interplay
Gene-environment interplay is a term encompassing multiple ways that genes and environments work together to produce a phenotype, or observable trait. Processes classified as examples of gene-environment interplay include gene-environment interaction, gene-environment correlation,[1] and epigenetics, which is the study of the effect of the environment on gene expression.[2] It is often studied with behavioral genetic research designs like twin, family, and adoption studies.[1]
Aim
The aim of studying gene-environment interplay is to discover new mechanisms of disease and to describe the reasons for digression from the expected expression of genes.[3] Studying these interactions allows for the complete understanding of diseases that are inclusive of numerous discrete and interacting cellular processes, including cell signalling.[3] By better understanding the underlying mechanisms of such diseases, interventions can be designed to target individual factors within specific populations that minimize the prevalence of disease.[3]
Types of Gene-Environment Interplay
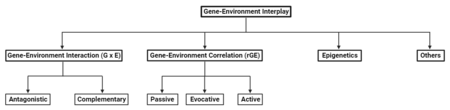
This is the most common form of gene-environment interplay. It occurs when genetic factors and environmental factors interact to produce an outcome that cannot be explained by either factor alone.[5] For example, a study found that individuals carrying the genetic variant 5-HTT (the short copy) were at a higher risk of developing depression when exposed to adverse childhood experiences, whereas those with other genotypes (long copy) were less affected by childhood maltreatment.[6]
This occurs when an individual's genotype influences the environments they are exposed to.[4] There are three subtypes of gene-environment correlation:
Passive Gene-Environment Correlation
This happens when parents provide both genes and environments for their children.[4] For example, parents who are genetically predisposed to be musically talented may also provide a musical environment for their children.[7]
Evocative Gene-Environment Correlation
In this case, an individual's genetic traits elicit responses from others in their environment.[4] For instance, a study has found that interpersonal control between a mother and her child, is partially a function of that child’s genetic predisposition toward control.[8]
Active Gene-Environment Correlation
This occurs when individuals seek out environments that are compatible with their genetic predispositions.[4] For example, a person with a genetic predisposition for athleticism may be more inclined to choose sports-related activities and environments.[9]
Environmental Factors
Pollutants
Epigenetic modifications can affect gene activity without changing the DNA sequence.[10] Decreased DNA methylation, the addition of a methyl group to a cytosine nucleotide on DNA, is thought to be linked to air pollution exposure. The mechanism behind this is not completely understood, but it may involve the formation of reactive oxygen species. These species generate oxidative stress, which simulates a chain reaction of signals in cells.[11] This can lead to hydroxymethylation, which replaces the methyl group with a hydroxyl group. Hydroxymethylation alters gene expression, potentially causing diseases such as lung cancer.[12] This can ultimately lead to inflammation in regions such as the airways, which triggers asthma. Cellular processes can also be activated to increase cytokine expression and immune cells in the same regions.[11] Cytokines are signalling proteins that regulate the immune system and are necessary when the body responds to injury or disease.[13]
Malnutrition
Nutrition can affect gene expression, thereby altering phenotype. Fetal starvation has been linked to decreased DNA methylation levels, particularly on the IGF2 gene associated with insulin metabolism.[14] This can increase the risk for metabolic disorders and type II diabetes mellitus.[15] Studies on prenatal exposure to famine have discovered that malnutrition causes differential DNA methylation of genes associated with growth, development and metabolism, increasing the risk of adverse phenotypes such as obesity and high cholesterol later in life.[16]
Exercise
Physical activity increases telomerase activity, which elongates the ends of chromosomes to maintain chromosomal stability, and induces epigenetic modifications of specific genes.[17] For example, it has been shown to increase methylation of the ASC gene, which generally decreases with age. Methylation can compact a gene, decreasing the amount of protein produced from the gene and the ASC gene stimulates cytokine production. Thus, the expression of inflammatory cytokines decreases.[18] This suppression can help prevent the development of chronic inflammation and associated age-related diseases due to excess inflammatory cytokines.[18] However, these epigenetic modifications depend on the intensity and type of exercise and are reversible with the cessation of physical activity.[19]
Prenatal Environment
The maternal environment can have epigenetic effects on the developing fetus. For instance, alcohol consumed during pregnancy can cross from maternal blood to the placenta and into the fetal environment of the amniotic cavity, where it can induce epigenetic modifications on fetal DNA.[20] Mouse embryo cultures show that alcohol exposure during fetal development can contribute to changes in DNA methylation of genes involved in development, metabolism, and organization of DNA during brain development.[21] These alcohol-induced changes in DNA methylation during pregnancy contribute to the distinct set of traits seen in Fetal Alcohol Spectrum Disorder (FASD).[21] Other instances of prenatal environment impact on fetal epigenetic state include maternal folic acid, stress, and tobacco smoking during pregnancy.[22][23][24]
Early Life Stress
Early life stress includes parental absence, abuse, and lack of bonding. These stressors in early childhood are associated with epigenetic modifications of the Hypothalamic-Pituitary-Adrenal (HPA) axis, which mediates stress response. Using a rat model of maternal care, reduced care between mother and offspring has been linked with down regulation of glucocorticoid receptors (GR) in the hypothalamus.[25] GRs are an important part of the HPA axis as they help restore normal physiological state after stress exposure. Down regulation of GRs expression occurs through histone modifications and DNA methylation of the GR gene, resulting in dysregulation of the stress response, including prolonged inflammation and cellular damage.[25] Several studies have also associated early life stress with later-life psychiatric disorders including anxiety and depression through epigenetic modulation of genes involved in the HPA axis.[26]
Studying Gene-Environment Interplay
Adoption and Twin Studies
Adoption and twin studies are used to investigate the complex interplay between genes and the environment. These studies typically involve the comparison of identical (monozygotic) and fraternal (dizygotic) twins to determine the extent to which genetic factors and environmental influences contribute to variations in traits or behaviors. These studies have contributed to studies of behaviour, personality, and psychiatric illnesses.[27] For example, a Finnish adoption study on schizophrenia revealed that a healthy environment can mitigate the effects of genetics in adopted individuals born to schizophrenic mothers.[28] Criminal and antisocial behaviour have also been found to be influenced by both genetic and environmental factors through these types of studies.[29][30]
Animal Models
Animal models provide a controlled and manipulable environment in which researchers can investigate the complex interactions between genes and environmental factors, shedding light on various biological and behavioural outcomes. For example, one study has demonstrated the utility of mouse models in understanding gene-environment interactions in schizophrenia due to the genetic similarities.[31]
Medical Conditions
Gene-environment interplay has been found to play a part in the majority of diseases. For instance, gene-environment interactions have a prevalent role in mental health disorders; specifically, evidence has found a link to alcohol dependence,[30] schizophrenia,[32] and psychosis.[33] There is a common polymorphism, or variant, in the AKT1 gene that causes its carriers who regularly use cannabis to be more susceptible to developing psychosis.[33] Evidence also supports gene-environment interplay to be connected to cardiovascular and metabolic conditions.[3] These include roles in obesity,[34] pulmonary disease,[35] and diabetes.[36] The rise in the incidence of diabetes is suggested to be linked to interactions between the FTO or KCNQ1 genes and environmental factors.[36]
References
- ↑ 1.0 1.1 Neiderhiser, Jenae M.; Liu, Chang; Griffin, Amanda M. (2018). "Gene–Environment Interplay". in Bornstein, Marc H.. The SAGE Encyclopedia of Lifespan Human Development. 2455 Teller Road, Thousand Oaks, California 91320: SAGE Publications, Inc.. pp. 939–940. doi:10.4135/9781506307633.n349. ISBN 9781506307657. http://sk.sagepub.com/reference/the-sage-encyclopedia-of-lifespan-human-development/i11932.xml.
- ↑ Rutter, Michael; Moffitt, Terrie E.; Caspi, Avshalom (9 November 2005). "Gene–environment interplay and psychopathology: multiple varieties but real effects" (in en). Journal of Child Psychology and Psychiatry 47 (3–4): 226–261. doi:10.1111/j.1469-7610.2005.01557.x. ISSN 0021-9630. PMID 16492258. https://acamh.onlinelibrary.wiley.com/doi/10.1111/j.1469-7610.2005.01557.x.
- ↑ 3.0 3.1 3.2 3.3 3.4 Flowers, Elena; Froelicher, Erika Sivarajan; Aouizerat, Bradley E. (2012). "Gene-environment interactions in cardiovascular disease". European Journal of Cardiovascular Nursing 11 (4): 472–478. doi:10.1016/j.ejcnurse.2011.06.001.
- ↑ 4.0 4.1 4.2 4.3 4.4 Jaffee, Sara R.; Price, Thomas S. (2008-12-01). "Genotype–environment correlations: implications for determining the relationship between environmental exposures and psychiatric illness". Psychiatry. Interplay between genes and environment in psychiatry 7 (12): 496–499. doi:10.1016/j.mppsy.2008.10.002. ISSN 1476-1793. PMID 20622930.
- ↑ Calam, C. T., ed. (1987), "Submerged culture conditions: the interaction between environment and genotype", Process Development in Antibiotic Fermentations, Cambridge Studies in Biotechnology (Cambridge: Cambridge University Press): pp. 65–74, doi:10.1017/cbo9780511983702.007, ISBN 978-0-521-30490-0, https://www.cambridge.org/core/books/process-development-in-antibiotic-fermentations/submerged-culture-conditions-the-interaction-between-environment-and-genotype/7343242893F1D7128817016013AF2900, retrieved 2023-12-04
- ↑ Caspi, Avshalom; Sugden, Karen; Moffitt, Terrie E.; Taylor, Alan; Craig, Ian W.; Harrington, HonaLee; McClay, Joseph; Mill, Jonathan et al. (2003-07-18). "Influence of Life Stress on Depression: Moderation by a Polymorphism in the 5-HTT Gene" (in en). Science 301 (5631): 386–389. doi:10.1126/science.1083968. ISSN 0036-8075. PMID 12869766. Bibcode: 2003Sci...301..386C. https://www.science.org/doi/10.1126/science.1083968.
- ↑ Corrigall, Kathleen A.; Schellenberg, E. Glenn (2015). "Predicting who takes music lessons: parent and child characteristics". Frontiers in Psychology 6: 282. doi:10.3389/fpsyg.2015.00282. ISSN 1664-1078. PMID 25852601.
- ↑ Klahr, Ashlea M.; Thomas, Katherine M.; Hopwood, Christopher J.; Klump, Kelly L.; Burt, S. Alexandra (February 2013). "Evocative gene–environment correlation in the mother–child relationship: A twin study of interpersonal processes" (in en). Development and Psychopathology 25 (1): 105–118. doi:10.1017/S0954579412000934. ISSN 0954-5794. PMID 23398756.
- ↑ Varillas-Delgado, David; Del Coso, Juan; Gutiérrez-Hellín, Jorge; Aguilar-Navarro, Millán; Muñoz, Alejandro; Maestro, Antonio; Morencos, Esther (2022-08-01). "Genetics and sports performance: the present and future in the identification of talent for sports based on DNA testing" (in en). European Journal of Applied Physiology 122 (8): 1811–1830. doi:10.1007/s00421-022-04945-z. ISSN 1439-6327. PMID 35428907.
- ↑ Weinhold, Bob (2006). "Epigenetics: The Science of Change" (in en). Environmental Health Perspectives 114 (3): A160–A167. doi:10.1289/ehp.114-a160. ISSN 0091-6765. PMID 16507447.
- ↑ 11.0 11.1 Rider, Christopher F.; Carlsten, Chris (2019-09-03). "Air pollution and DNA methylation: effects of exposure in humans". Clinical Epigenetics 11 (1): 131. doi:10.1186/s13148-019-0713-2. ISSN 1868-7083. PMID 31481107.
- ↑ Besaratinia, Ahmad; Caceres, Amanda; Tommasi, Stella (March 2022). "DNA Hydroxymethylation in Smoking-Associated Cancers". International Journal of Molecular Sciences 23 (5): 2657. doi:10.3390/ijms23052657. PMID 35269796.
- ↑ Foster, John (June 2001). "The functions of cytokines and their uses in toxicology". International Journal of Experimental Pathology 82 (3): 171–192. doi:10.1111/j.1365-2613.2001.iep192.x. PMID 11488991.
- ↑ Heijmans, Bastiaan T.; Tobi, Elmar W.; Stein, Aryeh D.; Putter, Hein; Blauw, Gerard J.; Susser, Ezra S.; Slagboom, P. Eline; Lumey, L. H. (2008-11-04). "Persistent epigenetic differences associated with prenatal exposure to famine in humans" (in en). Proceedings of the National Academy of Sciences 105 (44): 17046–17049. doi:10.1073/pnas.0806560105. ISSN 0027-8424. PMID 18955703. Bibcode: 2008PNAS..10517046H.
- ↑ Tiffon, Céline (2018). "The Impact of Nutrition and Environmental Epigenetics on Human Health and Disease" (in en). International Journal of Molecular Sciences 19 (11): 3425. doi:10.3390/ijms19113425. ISSN 1422-0067. PMID 30388784.
- ↑ Tobi, Elmar W; Slieker, Roderick C; Stein, Aryeh D; Suchiman, H Eka D; Slagboom, P Eline; van Zwet, Erik W; Heijmans, Bastiaan T; Lumey, LH (5 May 2015). "Early gestation as the critical time-window for changes in the prenatal environment to affect the adult human blood methylome". International Journal of Epidemiology 44 (4): 1211–1223. doi:10.1093/ije/dyv043. ISSN 0300-5771. PMID 25944819.
- ↑ Werner, Christian M; Hecksteden, Anne; Morsch, Arne; Zundler, Joachim; Wegmann, Melissa; Kratzsch, Jürgen; Thiery, Joachim; Hohl, Mathias et al. (1 January 2019). "Differential effects of endurance, interval, and resistance training on telomerase activity and telomere length in a randomized, controlled study". European Heart Journal 40 (1): 34–46. doi:10.1093/eurheartj/ehy585. ISSN 0195-668X. PMID 30496493. PMC 6312574. https://academic.oup.com/eurheartj/article/40/1/34/5193508.
- ↑ 18.0 18.1 Nakajima, K.; Takeoka, M.; Mori, M.; Hashimoto, S.; Sakurai, A.; Nose, H.; Higuchi, K.; Itano, N. et al. (3 March 2010). "Exercise Effects on Methylation of ASC Gene". International Journal of Sports Medicine 31 (9): 671–675. doi:10.1055/s-0029-1246140. ISSN 0172-4622. PMID 20200803. http://www.thieme-connect.de/DOI/DOI?10.1055/s-0029-1246140.
- ↑ Sellami, Maha; Bragazzi, Nicola; Prince, Mohammad Shoaib; Denham, Joshua; Elrayess, Mohamed (2021). "Regular, Intense Exercise Training as a Healthy Aging Lifestyle Strategy: Preventing DNA Damage, Telomere Shortening and Adverse DNA Methylation Changes Over a Lifetime". Frontiers in Genetics 12. doi:10.3389/fgene.2021.652497. ISSN 1664-8021. PMID 34421981.
- ↑ Gupta, Keshav K.; Gupta, Vinay K.; Shirasaka, Tomohiro (4 July 2016). "An Update on Fetal Alcohol Syndrome-Pathogenesis, Risks, and Treatment". Alcoholism: Clinical and Experimental Research 40 (8): 1594–1602. doi:10.1111/acer.13135. ISSN 1530-0277. PMID 27375266. https://onlinelibrary.wiley.com/doi/10.1111/acer.13135.
- ↑ 21.0 21.1 Liu, Yunlong; Balaraman, Yokesh; Wang, Guohua; Nephew, Kenneth P.; Zhou, Feng C. (22 May 2009). "Alcohol exposure alters DNA methylation profiles in mouse embryos at early neurulation" (in en). Epigenetics 4 (7): 500–511. doi:10.4161/epi.4.7.9925. ISSN 1559-2294. PMID 20009564.
- ↑ Crider, Krista S.; Yang, Thomas P.; Berry, Robert J; Bailey, Lynn B. (January 2012). "Folate and DNA Methylation: A Review of Molecular Mechanisms and the Evidence for Folate's Role" (in en). Advances in Nutrition 3 (1): 21–38. doi:10.3945/an.111.000992. PMID 22332098.
- ↑ Cao-Lei, L.; de Rooij, S. R.; King, S.; Matthews, S. G.; Metz, G. A. S.; Roseboom, T. J.; Szyf, M. (October 2020). "Prenatal stress and epigenetics". Neuroscience & Biobehavioral Reviews. Prenatal Stress and Brain Disorders in Later Life 117: 198–210. doi:10.1016/j.neubiorev.2017.05.016. ISSN 0149-7634. PMID 28528960. https://www.sciencedirect.com/science/article/pii/S0149763416307266.
- ↑ Nakamura, Aurélie; François, Olivier; Lepeule, Johanna (29 March 2021). "Epigenetic Alterations of Maternal Tobacco Smoking during Pregnancy: A Narrative Review" (in en). International Journal of Environmental Research and Public Health 18 (10): 5083. doi:10.3390/ijerph18105083. ISSN 1660-4601. PMID 34064931.
- ↑ 25.0 25.1 Liu, Dong; Diorio, Josie; Tannenbaum, Beth; Caldji, Christian; Francis, Darlene; Freedman, Alison; Sharma, Shakti; Pearson, Deborah et al. (12 September 1997). "Maternal Care, Hippocampal Glucocorticoid Receptors, and Hypothalamic-Pituitary-Adrenal Responses to Stress" (in en). Science 277 (5332): 1659–1662. doi:10.1126/science.277.5332.1659. ISSN 0036-8075. PMID 9287218. https://www.science.org/doi/10.1126/science.277.5332.1659.
- ↑ Wang, Fushun; Pan, Fang; Tang, Yiyuan; Huang, Jason H. (15 July 2021). "Editorial: Early Life Stress-Induced Epigenetic Changes Involved in Mental Disorders". Frontiers in Genetics 12. doi:10.3389/fgene.2021.684844. ISSN 1664-8021. PMID 34335692.
- ↑ Atkinson, Breanna E.; Vernon, Philip A. (18 September 2020), Carducci, Bernardo J.; Nave, Christopher S.; Nave, Christopher S., eds. (in en), Gene–Environment Interaction (1 ed.), Wiley, pp. 207–210, doi:10.1002/9781118970843.ch212, ISBN 978-1-118-97074-4, https://onlinelibrary.wiley.com/doi/10.1002/9781118970843.ch212, retrieved 2023-10-03
- ↑ Tienari, Pekka; Wynne, Lyman C.; Moring, Juha; Lahti, Ilpo; Naarala, Mikko; Sorri, Anneli; Wahlberg, Karl-Erik; Saarento, Outi et al. (April 1994). "The Finnish Adoptive Family Study of Schizophrenia: Implications for Family Research" (in en). The British Journal of Psychiatry 164 (S23): 20–26. doi:10.1192/S0007125000292696. ISSN 0007-1250. https://www.cambridge.org/core/journals/the-british-journal-of-psychiatry/article/abs/finnish-adoptive-family-study-of-schizophrenia/03CD46D224572B7E9A992863FA14B229.
- ↑ Cadoret, Remi J.; Cain, Colleen A.; Crowe, Raymond R. (May 1983). "Evidence for gene-environment interaction in the development of adolescent antisocial behavior". Behavior Genetics 13 (3): 301–310. doi:10.1007/BF01071875. ISSN 1573-3297. PMID 6615382. https://doi.org/10.1007/BF01071875.
- ↑ 30.0 30.1 van der Zwaluw, Carmen S.; Engels, Rutger C. M. E. (6 May 2009). "Gene-environment interactions and alcohol use and dependence: current status and future challenges" (in en). Addiction 104 (6): 907–914. doi:10.1111/j.1360-0443.2009.02563.x. ISSN 0965-2140. PMID 19466917. https://onlinelibrary.wiley.com/doi/10.1111/j.1360-0443.2009.02563.x.
- ↑ Ayhan, Yavuz; Sawa, Akira; Ross, Christopher A.; Pletnikov, Mikhail V. (2009-12-07). "Animal models of gene–environment interactions in schizophrenia". Behavioural Brain Research. Special Issue on Modeling Schizophrenia 204 (2): 274–281. doi:10.1016/j.bbr.2009.04.010. ISSN 0166-4328. PMID 19379776.
- ↑ van Os, Jim; Rutten, Bart PF; Poulton, Richie (6 November 2008). "Gene-Environment Interactions in Schizophrenia: Review of Epidemiological Findings and Future Directions". pp. 1066–1082. doi:10.1093/schbul/sbn117. https://academic.oup.com/schizophreniabulletin/article-lookup/doi/10.1093/schbul/sbn117.
- ↑ 33.0 33.1 Zwicker, Alyson; Denovan-Wright, Eileen M.; Uher, Rudolf (September 2018). "Gene-environment interplay in the etiology of psychosis" (in en). Psychological Medicine 48 (12): 1925–1936. doi:10.1017/S003329171700383X. ISSN 0033-2917. PMID 29334044. https://www.cambridge.org/core/journals/psychological-medicine/article/abs/geneenvironment-interplay-in-the-etiology-of-psychosis/249CDAFFD0387BE098046AF9F6B27F62.
- ↑ Andreasen, Camilla H.; Andersen, Gitte (2009-10-01). "Gene–environment interactions and obesity—Further aspects of genomewide association studies". Nutrition 25 (10): 998–1003. doi:10.1016/j.nut.2009.06.001. ISSN 0899-9007. PMID 19596186. https://www.sciencedirect.com/science/article/pii/S0899900709002469.
- ↑ Hirvonen, Ari (2009-07-10). "Gene–environment interactions in chronic pulmonary diseases". Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. Gene-environment interactions in noncancer degenerative diseases 667 (1): 132–141. doi:10.1016/j.mrfmmm.2008.12.013. ISSN 0027-5107. PMID 19563928. https://www.sciencedirect.com/science/article/pii/S0027510709000268.
- ↑ 36.0 36.1 Kido, Yoshiaki (2017-03-01). "Gene–environment interaction in type 2 diabetes" (in en). Diabetology International 8 (1): 7–13. doi:10.1007/s13340-016-0299-2. ISSN 2190-1686. PMID 30603301.
