Biology:Haplogroup I-M170
| Haplogroup I-M170 | |
|---|---|
| Possible time of origin | ~42,900 Years BP [2] |
| Coalescence age | ~27,500 Years BP [3] |
| Possible place of origin | Europe |
| Ancestor | IJ |
| Descendants | I*, I1, I2 |
| Defining mutations | L41, M170, M258, P19_1, P19_2, P19_3, P19_4, P19_5, P38, P212, U179 |
Haplogroup I (M170) is a Y-chromosome DNA haplogroup. It is a subgroup of haplogroup IJ, which itself is a derivative of the haplogroup IJK. Subclades I1 and I2 can be found in most present-day European populations, with peaks in some Northern European and Southeastern European countries.
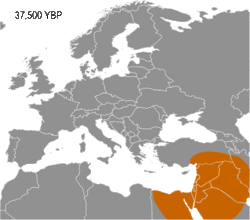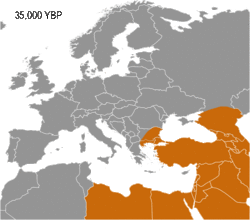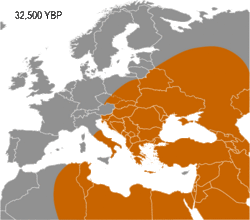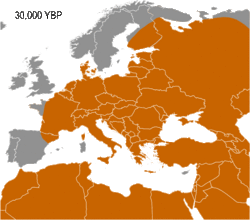
Haplogroup I appears to have arisen in Europe, so far being found in Palaeolithic sites throughout Europe (Fu 2016), but not outside it. It diverged from common ancestor IJ* about 43,000 years B.P. (Karafet 2008). Early evidence for haplogroup J has been found in the Caucasus and Iran (Jones 2015, Fu 2016). In addition, living examples of the precursor Haplogroup IJ* have been found only in Iran, among the Mazandarani and ethnic Persians from Fars.[1] This may indicate that IJ originated in South West Asia.
Haplogroup I has been found in multiple individuals belonging to the Gravettian culture. The Gravettians expanded westwards from the far corner of Eastern Europe, likely Russia, to Central Europe. They are associated with a genetic cluster that is normally called the Věstonice cluster.[2][3][4]
Origins
Available evidence suggests that I-M170 was preceded into areas in which it would later become dominant by haplogroups K2a (K-M2308) and C1 (Haplogroup C-F3393). K2a and C1 have been found in the oldest sequenced male remains from Western Eurasia (dating from circa 45,000 to 35,000 years BP), such as: Ust'-Ishim man (modern west Siberia) K2a*, Oase 1 (Romania) K2a*, Kostenki 14 (south west Russia) C1b, and Goyet Q116-1 (Belgium) C1a.[5][6] The oldest I-M170 found is that of an individual known as Krems WA3 (lower Austria), dating from circa 33,000-24,000 BP. At the same site, two twin boys were also found, both were assigned to haplogroup I*.[7][8]
Haplogroup IJ was in the Middle East and/or Europe about 40,000 years ago.[citation needed] The TMRCA (time to most recent common ancestor) for I-M170 was estimated by Karafet and colleagues in 2008 to be 22,200 years ago, with a confidence interval between 15,300 and 30,000 years ago.[9] This would make the founding event of I-M170 approximately contemporaneous with the Last Glacial Maximum (LGM), which lasted from 26,500 years ago until approximately 19,500 years ago.[10] TMRCA is an estimate of the time of subclade divergence. Rootsi and colleagues in 2004 also note two other dates for a clade, age of STR variation, and time since population divergence. These last two dates are roughly associated, and occur somewhat after subclade divergence. For Haplogroup I-M170 they estimate time to STR variation as 24,000 ±7,100 years ago and time to population divergence as 23,000 ±7,700 years ago.[11] These estimates are consistent with those of Karafet 2008 cited above. However, Underhill and his colleagues calculate the time to subclade divergence of I1 and I2 to be 28,400 ±5,100 years ago, although they calculate the STR variation age of I1 at only 8,100 ±1,500 years ago.[12]
Semino (2000) speculated that the initial dispersion of this population corresponds to the diffusion of the Gravettian culture.[13] Later the haplogroup, along with two cases of Haplogroup C, was found in human remains belonging to the previously mentioned Gravettian culture and in individuals of the Magdalenian and Azilian cultures.[14] Rootsi and colleagues in 2004 suggested that each of the ancestral populations now dominated by a particular subclade of Haplogroup I-M170 experienced an independent population expansion immediately after the Last Glacial Maximum.[11]
The five known cases of Haplogroup I from Upper Paleolithic European human remains make it one of the most frequent haplogroup from that period.[14] In 2016, the 31,210–34,580-year-old remains of a hunter-gatherer from Paglicci Cave, Apulia, Italy were found to carry I-M170.[15] So far, only Haplogroup F* and Haplogroup C1b have been documented, once each, on older remains in Europe. I2 subclade of I-M170 is the main haplogroup found on male remains in Mesolithic Europe, until circa 6,000 BCE, when mass migration into Europe of Anatolian farmers carrying Y-DNA G2a happened.[16]
Due to the arrival of so-called Early European Farmers (EEFs), I-M170 is outnumbered by Haplogroup G among Neolithic European remains and by Haplogroup R in later remains.
The earliest documentation of I1 is from Neolithic Hungary, although it must have separated from I2 at an earlier point in time.
In one instance, haplogroup I was found far from Europe, among 2,000-year-old remains from Mongolia.[17]
It would seem to be that separate waves of population movement impacted Southeastern Europe. The role of the Balkans as a long-standing corridor to Europe from Anatolia and/or the Caucasus is shown by the common phylogenetic origins of both haplogroups I and J in the parent haplogroup IJ (M429). This common ancestry suggests that the subclades of IJ entered the Balkans from Anatolia or the Caucasus, some time before the Last Glacial Maximum. I and J were subsequently distributed in Asia and Europe in a disjunctive phylogeographic pattern typical of "sibling" haplogroups. A natural geographical corridor like the Balkans is likely to have been used later by members of other subclades of IJ, as well as other haplogroups, including those associated with Early European Farmers.
The existence of Haplogroup IJK – the ancestor of both haplogroups IJ and K (M9) – and its evolutionary distance from other subclades of Haplogroup F (M89), supports the inference that haplogroups IJ and K both arose in Southwestern Asia. Living carriers of F* and IJ* have been reported from the Iranian Plateau.[1]
Distribution
Frequencies of Haplogroup I:
| Population | % hg I | % hg I (Subpopulation) | Sampled individuals | Source |
| Abazinians | 3.4 | 88 | Sergeevich 2007[18] | |
| Abkhazians | 33.3 | 12 | Nasidze Ivan 2004[19] | |
| Adyghe (Adygea) | 7 | 154 | [20] | |
| Adyghe (Cherkessia) | 2 | 126 | Sergeevich 2007[18] | |
| Adyghe (Kabardia) | 10 | 59 | Nasidze Ivan 2004[19] | |
| Afghanistan | 3 (Hazara people) | 60 | El Sibai 2009[21] | |
| Afghanistan | 1.5% | 3.3% (2/60) Hazara, 1.8% (1/56) Tajik | 204 | Haber et al. 2012[22] |
| Afghanistan | 0.99% | 2.6% (2/77) Hazara, 2.1% (3/142) Tajiks, 0/74 Turkmens, 0/87 Pashtuns, 0/127 Uzbeks | 507 | Di Cristofaro 2013[23] |
| Albanians | 13% (29/223) (Albania) | 223 | Sarno 2015 | |
| Albanians | 16 (Tosk), 4 (Gheg) | Ferri 2010 | ||
| Albanians | 21.82% (12/55) (Tirana) | 55 | Battaglia 2008 | |
| Albanians | 7 (Tirana) | 30 | Bosch 2006 | |
| Algerians | 0 | 156 | [24] | |
| Andis | 27 | |||
| Armenians | 5 | FTDNA 2013[25] | ||
| Avars | 2 | 115 | Balanovsky | |
| Austrians | 28 | 50 (Vienna), 29 (Graz), 6 (Tyrol) | [26] | |
| Ashkenazi | 1 | 1099 | [27] | |
| Azeri | 3 | 72 | Nasidze Ivan 2004 | |
| Balkars | 3 | 135 | Kutuev 2007[28] | |
| Belarusians | 23 | 11 (West), 15 (North), 16 (East), 28 (Centre), 30 (East Polesie), 34 (West Polesie) | 565 | Kushniarevich 2013 |
| Belarusians | 32 | Polesie- 43 (Vichin), 12 (Avtyuki) | 204 | Sergeevich 2015[29] |
| Bosnia and Herzegovina | 53 | 73 (Croats), 49 (Bosniaks), 33 (Serbs) | 256 | Marjanovic 2006 |
| Bosnia and Herzegovina | 65 | Herzegovina- 71 (Mostar, Siroki Brijeg), Bosnia- 54 (Zenica) | 210 | Pericic 2005[30] |
| Bosnia and Herzegovina | 73 (Croats), 45 (Bosniaks), 36 (Serbs) | 255 | Battaglia 2008[31] | |
| Bulgarians | 27-29 | 40 (Varna), 32 (Sofia), 30 (Plovdiv), 10 (Haskovo) | 935 | Karachanak 2009–13[32][33] |
| Bulgarians | 34 | 100 | Begona Martinez-Cruz 2012 | |
| Bulgaria | 19 (Bulgarian Turks) | 63 | Zaharova 2002[34] | |
| Central Asia | 2 | 984 | Rootsi 2004 | |
| Chechens | 0 | 330 | Balanovsky | |
| Croats | 45 | 1100 | Mrsic 2012 | |
| Croats | 47 | 55 (Hvar), 52 (Osijek), 41 (Pula), 57 (Split), 29 (Varaždin) | 518 | Primorac 2022[35] |
| Cyprus | 1 | 164 | El-Sibai 2009[36] | |
| Czechs | 18 | 25 (Klatovy), 25 (Písek), 15 (Brno) 14 (Hradec Králové), 10 (Třebíč) | 257 | Luca 2007[37] |
| Danes | 49 | 194 | Rootsi 2004 | |
| Darginians | 58 | 26 | Nasidze Ivan 2004 | |
| Darginians (Kaitak) | 0 | 101 | ||
| Dutch | 27.8 | 2085 | Altena 2020[38] | |
| Dutch | 33 | 410 | Van Doorn 2008[39] | |
| Egyptians | 0 | 124 | El-Sibai 2009[40] | |
| Egyptians | 1 | 370 | [24] | |
| Estonians | 19 | 194 | Rootsi 2004 | |
| English | 18 | 945 | Rootsi 2004 | |
| English | 26 | 12 (Cornwall), 38 (Essex) | 1830 | FTDNA 2016[41] |
| Estonians | 17 | 118 | Lappalainen 2008[42] | |
| Flemish Belgians | 28 | 113 | [43] | |
| Finland | 29 | 36 (Swedes from Ostrobothnia), 15 (Northern Savo) | 536 | Lappalainen 2006[44] |
| French | 16 (South), 24 (Normandy), 4 (Lyon), 4 (Corsica) | Rootsi 2004 | ||
| French | 9 | 5 (Auvergne), 13 (Brittany), 9 (Nord Pas de Calais) | 555 | Ramos-Luis 2009[45] |
| French | 13 | 11 (Paris), 18 (Strasburg), 10 (Lyon) | 333 | Kari Hauhio[26] |
| Gagauzes | 28 | 89 | Varzari 2006 | |
| Georgians | 0 | 63 | Rootsi 2004 | |
| Georgians | 4 | 77 | Nasidze Ivan 2004 | |
| Germans | 24 | 32 (Berlin), 32 (Hamburg), 15 (Leipzig) | 1215 | Kayser 2005[46] |
| Greeks | 14 | 30 (Macedonia) | 261 | Rootsi 2004 |
| Greeks | 10 (Athens), 30 (Macedonia) | 149 | Battaglia 2008 | |
| Greeks | 36 (Serres), 24 (Agrinio), 20 (Thessaloniki), 18 (Mytilene), 14 (Crete), 14 (Larissa), 11 (Patrai), 12 (Karditsa), 8 (Ioannina), 2 (Chios) | 366 | Di Giacommo 2003[47] | |
| Greeks | 12 (North), 24 (South) | 142 | Zalloua 2008 | |
| Greenlanders | 17 | 215 | Sanchez 2004[48] | |
| Hungarians | 23 | 162 | Rootsi 2004 | |
| Hungarians | 28 | 230 | Vago Zalan Andrea 2008 | |
| Indians | 0 (North India) | 560 | [49] | |
| Ingush | 0 | 143 | ||
| Iranians | 2 | 22 (South Iran), 5 (Khorasan), 0 (Teheran) | 186 | Di Cristofaro 2013[23] |
| Iranians | 1 (West), 1 (East) | 324 | [50] | |
| Iranians | 0 | 83 | Rootsi 2004 | |
| Iranians | 1 | 92 | El-Sibai 2009[36] | |
| Iranians | 0 | 6 (Armenians of Teheran), 0 (Persians of Teheran, Fars, Isfahan, Khorasan, Yazd) | 952 | Grugni 2012 |
| Iraqis | 1 | 176 | Rootsi 2004[51] | |
| Iraqis | 1 | 117 | El-Sibai 2009[36] | |
| Irish | 11 | 76 | Rootsi 2004 | |
| Irish | 10 | 119 | Cappeli 2013[52] | |
| Irish | 11 (Rush, Dublin) | Capelli 2003 | ||
| Italians | 5 (North), 7 (Central), 9 (South and Sicily), 39 (Sardinia) | Rootsi 2004 | ||
| Italians | 10 | 31 (Sardinia), 4 (Umbria, Marche) | 884 | Boattini 2013[53] |
| Italians | 7 | 0 (Calabria, Pescara, Garfagnana, Val di Non), 5 (Verona), 7(Genoa), 19 (Foggia) | 524 | Di Giacomo 2003[54] |
| Italians | 36 (Filettino) 35 (Cappadocia, Abruzzo), 28 (Vallepietra) | Messina 2015[55] | ||
| Italians | 23 (Udine), 17 (Saniti), 13 (Picenium), 7 (Latini) | 583 | Brisighelli 2012[56] | |
| Italians | 30 (Stelvio) | [57] | ||
| Italians | 31 (Caccamo) | Gaetano 2008[58] | ||
| Jordanians | 1 | 273 | El-Sibai 2009[36] | |
| Jordanians | 5 (Amman), 0 (Dead Sea) | 146 | Flores 2005[59] | |
| Kara Nogays | 13 | 76 | ||
| Karachays | 9 | 69 | Sergeevich 2007[18] | |
| Kazakhs | 1 | 370 | [60] | |
| Kosovar Albanians | 8 | 114 | Pericic 2005 | |
| Kumyks | 0 | 73 | Kutuev 2007[28] | |
| Kurds | 4 (West Iran) | 21 | Malyarchuk 2013[61] | |
| Kurds | 2 (Iran) | 59 | Gragni 2012 | |
| Kurmanji | 17 (Turkey), 0 (Georgia) | 112 | Nasidze 2005[62] | |
| Kuwaiti | 0 | 42 | El-Sibai 2009[36] | |
| Kyrgyzstan | 0 (Uyghurs), 0 (Kyrgyz) | Di Cristofaro 2013[23] | ||
| Laks | 14 | [63] | ||
| Latvians | 9 | 3 (Southwest) | [64] | |
| Lebanese | 3 | 10 (North Maronite), 0 (Shia) | 951 | [36][65] |
| Lebanese | 5 | 66 | Rootsi 2004 | |
| Lezgis | 0 | 81 | ||
| Lithuanians | 7 | Kushniarevich 2015 | ||
| Libyans | 0 | 83 | [24] | |
| Libyans | 2 | 1 | 175 | Fendri 2015[66] |
| Macedonians | 34 (Skopje) | 79 | Pericic 2005 | |
| Macedonians | 24 | 31 (Macedonians), 12 (Albanians) | 343 | Noevski 2010 |
| Macedonians | 13 (Albanians) | 64 | Battaglia 2008[31] | |
| Maltese | 12 | 90 | El-Sibai 2009[36] | |
| Moldovans | 29 (Moldovans), 25 (Ukrainians) | Varzari 2006 | ||
| Moroccans | 0 | 316 | El-Sibai 2009[36] | |
| Moroccans | 0 | 760 | [24] | |
| Mongols | 1 | 160 | Di Cristofaro 2013[23] | |
| Norwegians | 37 | 40 (Oslo) 30 (West), 42 (East, South), 35 (North), 33 (Bergen) | Dupuy 2005 | |
| Pakistan | 0 | 638 | [67] | |
| Poles | 17 | 19 (Warsaw), 12 (Lublin), 22 (Szczecin) | 913 | Kayser 2005 |
| Poles | 18 | 191 | Rootsi 2004 | |
| Portuguese | 5 | 303 | Rootsi 2004 | |
| Portuguese | 8 | 3 (Lisboa), 0 (Setubal), 18 (Braga) | 657 | Beleza 2005[68] |
| Qatar | 0 | 72 | El-Sibai 2009[36] | |
| Romani | 17 (Hungary), 10 (Tiszavasvari), 5 (Tokaj) 37 (Taktakoz), 11 (Slovakia) | Vago Zalan Andrea 2008 | ||
| Romanians | 28 | 36 (Brasov), 18 (Cluj) | 178 | Martinez-Cruz 2012[69] |
| Romanians | 22 | 361 | Rootsi 2004 | |
| Russians | 13 (North Europe), 18 (Centre Europe), 21 (South Europe), 27 (Unzha), 0 (Mezen) | 1228 | Balanovsky 2008 | |
| Russia | 2 (Udmurts), 5 (Pinega), 5 (Komi), 5 (Tatars), 6 (Bashkortostan), 7 (Chuvashes), 19 (Kostroma), 11 (Smolensk), 17 (Belgorod), 19 (Mordvins), 23 (Cossacks), 24 (Adygea) | Rootsi 2004 | ||
| Saami | 31 | Rootsi 2004 | ||
| Saudis | 0 | 1597 | [70] | |
| Scotland | 11 | 17 (Scottish Isles) | Rootsi 2004 | |
| Sephardi | 4 (Portugal) | 57 | ||
| Serbs | 39 | Serbia with Kosovo | 209 | Zgonjanin 209[71] |
| Serbs | 48 (Serbia), 39 (Kosovo), 52 (Herzegovina and Montenegro) | 1200 | Mihajlovic 2022[72] | |
| Slovaks | 28 | 250 | Petrejcikova 2013[73] | |
| Slovenians | 30 | 57 (Spodnjeposavska) | 458 | Vakar 2010[74] |
| Spaniards | 6 | 18 (Asturias), 0 (Gascony) | 1002 | Adams 2008[75] |
| Sudanese | 5 (Nubians), 4 (Gaalien), 7 (Mesereia) | [76] | ||
| Swedes | 42 | 32 (Ostergotaland & Jonkoping) 50 (Gotland & Varmland) | 305 | Karlsson2006[77] |
| Swedes | 26 (North Sweden),[78] | Rootsi2004 | ||
| Swedes | 41 (South), 26 (North) | Rootsi 2004 | ||
| Swedes | 44 | 60 (Kristianstad), 60 (Kalmar), 59 (Kronoberg), 55 (Stockholm), 37 (North Norrland), 52 (South Norrland) | 1800 | FTDNA 2016[79] |
| Swiss | 8 | 144 | Rootsi 2004 | |
| Swiss | 23 | 13 (Lausanne), 32 (Bern) | [26] | |
| Syrians | 2 (West), 3 (East) | 520 | [50] | |
| Syrians | 2 | 554 | El Sibai 2009[36] | |
| Tataers | 33 (China) | 33 | [80] | |
| Tunisians | 0 | El-Sibai 2009[36] | ||
| Tunisians | 0 | 601 | [24] | |
| Turks | 5 | 12 (Marmara), 10 (Istanbul), 7 (Western Anatolia), 4 (Central Anatolia), 0 (Eastern Anatolia) | 523 | Cinnioglu 2003 |
| Turks | 5 | 741 | Rootsi 2004 | |
| UAE | 0 | 164 | El-Sibai 2009[36] | |
| Ukrainians | 22 | 585 | Rootsi 2004 | |
| Ukrainians | 28 | 33 (Sumy), 23 (Ivano-Frankivsk) | 701 | Kushniarevich 2013 |
| Welsh | 8 | 196 | Rootsi 2004 | |
| Yemenese | 0 | 62 | El-Sibai 2009[36] | |
| Zazas | 33 (Turkey) | 27 | Nasidze 2005[62] | |
| 17 (Albanians in Tirana), 29 (Macedonians in Skopje), 21 (Aromanians in Krusevo), 19 (Greeks in Thrace), 42 (Aromanians in Andon Poci), 42 (Romanians in Constanta), 39 (Romanians in Piteşti) | Bosch 2006[81] | |||
| 47 (Romanians from Buhusi and Piatra Neamț), 35 (Moldovans from Sofia), 24 (Moldovans from Karasahani) 24 (Gagauzes from Etulia), 31 (Gagauzes from Kongaz), 25 (Ukrainians from Rashkovo) | Vazari 2006 | |||
| 38 (Sweden), 41 (Western Finland), 28 (Eastern Finland), 18 (Karelia), 12 (Lithuania), 7 (Latvia), 17 (Estonia) | Lappalainen2008[82] | |||
| 34 (Iranians from Teheran), 10 (Iranians from Isfahan), 32 (Ossetians from Ardon), 13 (Ossetians from Digora) | Nasidze Ivan. 2004[19] | |||
| 3 (Tajiks) 3 (East Persians) | Malyarchuk 2013[61] | |||
| 2 (Kizhi), 4 (Teleuts), 4 (Khakassians), 3 (Todjins), 2 (Evenks) 3 (Tofalars), 1 (Tuvinians) |
Subgroups
The subclades of Haplogroup I-M170 with their defining mutations, as of 2011.[83] Up-to-date phylogenetic trees listing all currently known subclades of I can be found at Y-Full and FamilyTreeDNA
- I-M170 ( L41, M170, M258, P19_1, P19_2, P19_3, P19_4, P19_5, P38, P212, Page123, U179) Middle East, Caucasus, Europe.
- I-M253 Haplogroup I1 (L64, L75, L80, L81, L118, L121/S62, L123, L124/S64, L125/S65, L157, L186, L187, M253, M307.2/P203.2, M450/S109, P30, P40, S63, S66, S107, S108, S110, S111) Typical of populations of Scandinavia and Northwest Europe, with a moderate distribution throughout Eastern Europe In Anatolia at 1%[84]
- I1a DF29/S438
- I1a1 CTS6364/Z2336
- I1a1a M227
- I1a1a1 M72
- I1a1b L22/S142
- I1a1b1 P109
- I1a1b2 L205
- I1a1b3 L287
- I1a1b3a L258/S335
- I1a1b3a1 L296
- I1a1b3a L258/S335
- I1a1b4 L300/S241
- I1a1b5 L813/Z719
- I1a1a M227
- I1a2 S244/Z58
- I1a2a S246/Z59
- I1a2a1 S337/Z60, S439/Z61, Z62
- I1a2a1a Z140, Z141
- I1a2a1a1 Z2535
- I1a2a1a1a L338
- I1a2a1a2 F2642
- I1a2a1a1 Z2535
- I1a2a1b Z73
- I1a2a1c L573
- I1a2a1d L1248
- I1a2a1d1 L803
- I1a2a1a Z140, Z141
- I1a2a2 Z382
- I1a2a1 S337/Z60, S439/Z61, Z62
- I1a2b S296/Z138, Z139
- I1a2b1 Z2541
- I1a2a S246/Z59
- I1a3 S243/Z63
- I1a3a L1237
- I1a1 CTS6364/Z2336
- I1b Z131[85]
- I1a DF29/S438
- I-M438 Haplogroup I2 L68/PF3781/S329, M438/P215/PF3853/S31
- I2a L460/PF3647/S238
- I2a1 P37.2
- I2a1a L158/PF4073/S433, L159.1/S169.1, M26/PF4056
- I2a1a1 L160/PF4013
- I2a1b L178/S328, M423
- I2a1b1 L161.1/S185
- I2a1b2 L621/S392
- I2a1b2a1a L147.2
- I2a1c L233/S183
- I2a1a L158/PF4073/S433, L159.1/S169.1, M26/PF4056
- I2a2 L35/PF3862/S150, L37/PF6900/S153, L181, M436/P214/PF3856/S33, P216/PF3855/S30, P217/PF3854/S23, P218/S32
- I2a2a L34/PF3857/S151, L36/S152, L59, L368, L622, M223, P219/PF3859/S24, P220/S119, P221/PF3858/S120, P222/PF3861/U250/S118, P223/PF3860/S117, Z77
- I2a2a1 CTS616, CTS9183
- I2a2a1a M284
- I2a2a1a1 L1195
- I2a2a1a1a L126/S165, L137/S166, L369
- I2a2a1a1b L1193
- I2a2a1a1 L1195
- I2a2a1b L701, L702
- I2a2a1b1 P78
- I2a2a1b2 L699, L703
- I2a2a1b2a L704
- I2a2a1c Z161
- I2a2a1c1 L801/S390
- I2a2a1c1a CTS1977
- I2a2a1c1a1 P95
- I2a2a1c1b CTS6433
- I2a2a1c1b1 Z78
- I2a2a1c1b1a L1198
- I2a2a1c1b1a1 Z190
- I2a2a1c1b1a1a S434/Z79
- I2a2a1c1b1a1 Z190
- I2a2a1c1b1a L1198
- I2a2a1c1b1 Z78
- I2a2a1c1a CTS1977
- I2a2a1c2 L623, L147.4
- I2a2a1c1 L801/S390
- I2a2a1d L1229
- I2a2a1d1 Z2054
- I2a2a1d1a L812/S391
- I2a2a1d2 L1230
- I2a2a1d1 Z2054
- I2a2a1a M284
- I2a2a2 L1228
- I2a2a1 CTS616, CTS9183
- I2a2b L38/S154, L39/S155, L40/S156, L65.1/S159.1, L272.3
- I2a2b1 L533
- I2a2a L34/PF3857/S151, L36/S152, L59, L368, L622, M223, P219/PF3859/S24, P220/S119, P221/PF3858/S120, P222/PF3861/U250/S118, P223/PF3860/S117, Z77
- I2a1 P37.2
- I2b L415, L416, L417
- I2c L596/PF6907/S292, L597/S333
- I2a L460/PF3647/S238
- I-M253 Haplogroup I1 (L64, L75, L80, L81, L118, L121/S62, L123, L124/S64, L125/S65, L157, L186, L187, M253, M307.2/P203.2, M450/S109, P30, P40, S63, S66, S107, S108, S110, S111) Typical of populations of Scandinavia and Northwest Europe, with a moderate distribution throughout Eastern Europe In Anatolia at 1%[84]
Note that the naming of some of the subgroups has changed, as new markers have been identified, and the sequence of mutations has become clearer.
I-M170
The composite subclade I-M170 contains individuals directly descended from the earliest members of Haplogroup I, bearing none of the subsequent mutations which identify the remaining named subclades.
Several I* individuals, who do not fall into any known subclades, have been found among the Lak people of Dagestan, at a rate of (3/21),[86] as well as Turkey (8/741), Adygea in the Caucasus (2/138) and Iraq (1/176), even though I-M170 occurs at only very low frequencies among modern populations of these regions as a whole. This is consistent with the belief that the haplogroup first appeared in South West Eurasia.
There are also high frequencies of Haplogroup I* among the Andalusians (3/103), French (4/179), Slovenians (2/55), Tabassarans (1/30),[86] and Saami (1/35).[87]
(Neither study from which the above figures were drawn excluded the present I2-M438 clade as a whole, but only certain subclades, so these presumed cases I* may possibly belong to I2.)
A living Hazara male from Afghanistan has also been found to carry I*, with all known subclades of both I1 (M253) and I2 (M438) ruled out.[88]
I1-M253
Haplogroup I1-M253 (M253, M307, P30, P40) displays a very clear frequency gradient, with a peak frequency of approximately 35% among the populations of southern Norway, southwestern Sweden, and Denmark, and rapidly decreasing frequencies toward the edges of the historically Germanic-influenced world. A notable exception is Finland, where frequency in West Finns is up to 40%, and in certain provinces like Satakunta more than 50%. I1 is believed to have become common as a result of a founder effect during the Nordic Bronze Age, and subsequently spread throughout Europe during the Migration Period when Germanic tribes migrated from southern Scandinavia and northern Germany to other places in Europe.[89]
Outside Fennoscandia, distribution of Haplogroup I1-M253 is closely correlated with that of Haplogroup I2a2-M436; but among Scandinavians (including both Germanic and Uralic peoples of the region) nearly all the Haplogroup I-M170 Y-chromosomes are I1-M253. Another characteristic of the Scandinavian I1-M253 Y-chromosomes is their rather low haplotype diversity (STR diversity): a greater variety of Haplogroup I1-M253 Y-chromosomes has been found among the French and Italians, despite the much lower overall frequency of Haplogroup I1-M253 among the modern French and Italian populations. This, along with the structure of the phylogenetic tree of I1-M253 strongly suggests that most living I1 males are the descendants of an initially small group of reproductively successful men who lived in Scandinavia during the Nordic Bronze Age.[90][91]
I2-M438
Haplogroup I2-M438, previously I1b, may have originated in southern Europe – it is now found at its highest frequencies in the western Balkans and Sardinia – some 15,000–17,000 years ago and developed into three main subgroups : I2-M438*, I2a-L460, I2b-L415 and I2c-L596.
I2a1a-M26
Haplogroup I2a1a-M26 is notable for its strong presence in Sardinia. Haplogroup I-M170 comprises approximately 40% of all patrilines among the Sardinians, and I2a1a-M26 is the predominant type of I among them.
Haplogroup I2a1a-M26 is practically absent east of France and Italy,[92] while it is found at low but significant frequencies outside of Sardinia in the Balearic Islands, Castile-León, the Basque Country, the Pyrenees, southern and western France, and parts of the Maghreb in North Africa, Great Britain, and Ireland. Haplogroup I2a1a-M26 appears to be the only subclade of Haplogroup I-M170 found among the Basques, but appears to be found at somewhat higher frequencies among the general populations of Castile-León in Spain and Béarn in France than among the population of ethnic Basques.[citation needed] The M26 mutation is found in native males inhabiting every geographic region where megaliths may be found, including such far-flung and culturally disconnected regions as the Canary Islands, the Balearic Isles, Corsica, Ireland, and Sweden.[92]
The distribution of I2a1a-M26 also mirrors that of the Atlantic Bronze Age cultures, which indicates a potential spread via the obsidian trade or a regular maritime exchange of some of metallurgical products.[92]
I2a1b-M423
Haplogroup I2a1b-M423 is the most frequent Y-chromosome haplogroup I-M170 in Central and Eastern European populations, reaching its peak in the Western Balkans, most notably in Dalmatia (50–60%[30]) and Bosnia-Herzegovina (up to 71%,[93] avg. 40-50%[30]). Its subclade I-L161 has greater variance in Ireland and Great Britain, but overall frequency is very low (2–3%), while subclade I-L162 has the highest variance and also high concentration in Eastern Europe (Ukraine, Southeastern Poland, Belarus).[94]
I2a2-M436
The distribution of Haplogroup I2a2-M436 (M436/P214/S33, P216/S30, P217/S23, P218/S32) is closely correlated to that of Haplogroup I1 except in Fennoscandia, which suggests that it was probably harbored by at least one of the Paleolithic refuge populations that also harbored Haplogroup I1-M253; the lack of correlation between the distributions of I1-M253 and I2a2-M436 in Fennoscandia may be a result of Haplogroup I2a2-M436's being more strongly affected in the earliest settlement of this region by founder effects and genetic drift due to its rarity, as Haplogroup I2a2-M436 comprises less than 10% of the total Y-chromosome diversity of all populations outside of Lower Saxony. Haplogroup I2a2-M436 has been found in over 4% of the population only in Germany, the Netherlands, Belgium, Denmark, England (not including Cornwall), Scotland, and the southern tips of Sweden and Norway in Northwest Europe; the provinces of Normandy, Maine, Anjou, and Perche in northwestern France; the province of Provence in southeastern France; the regions of Tuscany, Umbria, and Latium in Italy; and Moldavia and the area around Russia's Ryazan Oblast and Republic of Mordovia in Eastern Europe. One subclade of Haplogroup I2a2-M436, namely I2a2a1a1-M284, has been found almost exclusively among the population of Great Britain, which has been taken to suggest that the clade may have a very long history in that island. It is notable, however, that the distributions of Haplogroup I1-M253 and Haplogroup I2a2-M436 seem to correlate fairly well with the extent of historical influence of Germanic peoples. The punctual presence of both haplogroups at a low frequency in the area of the historical regions of Bithynia and Galatia in Turkey may be related to the Varangian Guard or rather suggests a connection with the ancient Gauls of Thrace, several tribes of which are recorded to have immigrated to those parts of Anatolia at the invitation of Nicomedes I of Bithynia. This suggestion is supported by recent genetic studies regarding Y-DNA Haplogroup I2b2-L38 have concluded that there was some Late Iron Age migration of Celtic La Tène people, through Belgium, to the British Isles including north-east Ireland.[95]
Haplogroup I2a2-M436 also occurs among approximately 1% of Sardinians, and in Hazaras from Afghanistan at 3%.[96]
Specifications of mutation
The technical details of U179 are:
- Nucleotide change (rs2319818): G to A
- Position (base pair): 275
- Total size (base pairs): 220
- Forward 5′→ 3′: aaggggatatgacgactgatt
- Reverse 5′→ 3′: cagctcctcttttcaactctca
Height
This haplogroup reaches its maximum frequency in the Western Balkans (with the highest concentration of I2 in present-day Herzegovina). It may be associated with unusually tall males, since those in the Dinaric Alps have been reported to be the tallest in the world, with an average male height of the range 180 cm (5 ft 11 in)–182 cm (6 ft 0 in) in the cantons of Bosnia, 184 cm (6 ft 0 in) in Sarajevo, 182 cm (6 ft 0 in)–186 cm (6 ft 1 in) in the cantons of Herzegovina mostly populated by Croats.[97] A 2014 study examining the correlation between Y-DNA haplogroups and height found a correlation between the haplogroups I1, R1b-U106, I2a1b-M423 and tall males.[98] The study featured the measured average heights of young German, Swedish, Dutch, Danish, Serbian and Bosnian men. The German male average height was 180.2 cm, the Swedish men were on average 181.4 cm, the Dutch men were 183.8 cm, the Danish men were 180.6 cm, the Serbians were 180.9 cm, and Bosnian Croat men from Herzegovina were 185.2 centimeters on average.
See also
- Haplogroup
- Human Y-chromosome DNA haplogroups
- Haplogroup I1 (Y-DNA)
- Haplogroup I2 (Y-DNA)
- Late Glacial Maximum
- Aurignacian
- Proto-Indo-Europeans
- Gravettian
Notes
- "Y chromosomal heritage of Croatian population and its island isolates". Eur. J. Hum. Genet. 11 (7): 535–42. July 2003. doi:10.1038/sj.ejhg.5200992. PMID 12825075. http://evolutsioon.ut.ee/publications/Barac2003.pdf.
- Bennett, E.A., Prat, S., Péan, S., Crépin, L., Yanevich, A., Puaud, S., ... & Geigl, E. M. (2019). The origin of the Gravettians: genomic evidence from a 36,000-year-old Eastern European. BioRxiv, 685404.
- "Phylogeography of Y-chromosome haplogroup I reveals distinct domains of prehistoric gene flow in Europe". Am. J. Hum. Genet. 75 (1): 128–137. 2019. doi:10.1086/422196. PMID 12825075.
- The Genographic Project, National Geographic, Atlas of the Human Journey
- ISOGG, Y-DNA Haplogroup I and its Subclades
References
- ↑ 1.0 1.1 Grugni (2012). "Ancient Migratory Events in the Middle East: New Clues from the Y-Chromosome Variation of Modern Iranians". PLOS ONE 7 (7): e41252. doi:10.1371/journal.pone.0041252. PMID 22815981. Bibcode: 2012PLoSO...741252G.
- ↑ "Publications Detail View" (in en). https://fgga.univie.ac.at/en/research/publications/publications-detail-view/?tx_univiepure_univiepure%5Buuid%5D=cb9edcf2-a5ca-469d-bd5b-c8021203aacc&tx_univiepure_univiepure%5Bwhat2show%5D=publ&tx_univiepure_univiepure%5Baction%5D=show&tx_univiepure_univiepure%5Bcontroller%5D=Pure&cHash=61ab4ac40430e51782aeea80a7bc7258.
- ↑ Mounier, Aurélien; Heuzé, Yann; Samsel, Mathilde; Vasilyev, Sergey; Klaric, Laurent; Villotte, Sébastien (2020-12-14). "Gravettian cranial morphology and human group affinities during the European Upper Palaeolithic" (in en). Scientific Reports 10 (1): 21931. doi:10.1038/s41598-020-78841-x. ISSN 2045-2322. PMID 33318530. Bibcode: 2020NatSR..1021931M.
- ↑ Bennett, E. Andrew; Prat, Sandrine; Péan, Stéphane; Crépin, Laurent; Yanevich, Alexandr; Puaud, Simon; Grange, Thierry; Geigl, Eva-Maria (2019-07-02). "The origin of the Gravettians: genomic evidence from a 36,000-year-old Eastern European" (in en). bioRxiv: 685404. doi:10.1101/685404. https://www.biorxiv.org/content/10.1101/685404v2.
- ↑ Fu, Qiaomei (2016). "The genetic history of Ice Age Europe". Nature 534 (7606): 200–5. doi:10.1038/nature17993. PMID 27135931. Bibcode: 2016Natur.534..200F.
- ↑ Seguin-Orlando et al. (2014)「Genomic structure in Europeans dating back at least 36,200 years」
- ↑ Teschler-Nicola, Maria; Fernandes, Daniel; Händel, Marc; Einwögerer, Thomas; Simon, Ulrich; Neugebauer-Maresch, Christine; Tangl, Stefan; Heimel, Patrick et al. (2020-11-06). "Ancient DNA reveals monozygotic newborn twins from the Upper Palaeolithic" (in en). Communications Biology 3 (1): 650. doi:10.1038/s42003-020-01372-8. ISSN 2399-3642. PMID 33159107.
- ↑ "Y-SNP calls for Krems WA3" (in en). 2016-05-11. https://genetiker.wordpress.com/y-snp-calls-for-krems-wa3/.
- ↑ "New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree". Genome Research 18 (5): 830–8. 2008. doi:10.1101/gr.7172008. PMID 18385274. PMC 2336805. http://www.genome.org/cgi/content/abstract/gr.7172008v1.
- ↑ "The Last Glacial Maximum". Science 325 (5941): 710–4. August 2009. doi:10.1126/science.1172873. PMID 19661421. Bibcode: 2009Sci...325..710C.
- ↑ 11.0 11.1 Rootsi SiiriExpression error: Unrecognized word "etal". (2004). "Phylogeography of Y-Chromosome Haplogroup I-M170 Reveals Distinct Domains of Prehistoric Gene Flow in Europe". American Journal of Human Genetics 75 (1): 128–137. doi:10.1086/422196. PMID 15162323.
- ↑ P.A. Underhill, N.M. Myres, S. Rootsi, C.T. Chow, A.A. Lin, R.P. Otillar, R. King, L.A. Zhivotovsky, O. Balanovsky, A. Pshenichnov, K.H. Ritchie, L.L. Cavalli-Sforza, T. Kivisild, R. Villems, S.R. Woodward, New Phylogenetic Relationships for Y-chromosome Haplogroup I: Reappraising its Phylogeography and Prehistory, in P. Mellars, K. Boyle, O. Bar-Yosef and C. Stringer (eds.), Rethinking the Human Evolution (2007), pp. 33–42.
- ↑ "The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective". Science 290 (5494): 1155–9. November 2000. doi:10.1126/science.290.5494.1155. PMID 11073453. Bibcode: 2000Sci...290.1155S.
- ↑ 14.0 14.1 "Palaeolithic DNA from Eurasia". http://www.ancestraljourneys.org/palaeolithicdna.shtml.
- ↑ Fu, Qiaomei; Posth, Cosimo; Hajdinjak, Mateja; Petr, Martin; Mallick, Swapan; Fernandes, Daniel; Furtwängler, Anja; Haak, Wolfgang et al. (2016). "The genetic history of Ice Age Europe" (in en). Nature 534 (7606): 200–205. doi:10.1038/nature17993. ISSN 0028-0836. PMID 27135931. Bibcode: 2016Natur.534..200F.
- ↑ "Ancient DNA reveals 'genetic continuity' between Stone Age and modern populations in East Asia". February 2017. http://www.cam.ac.uk/research/news/ancient-dna-reveals-genetic-continuity-between-stone-age-and-modern-populations-in-east-asia.
- ↑ Keyser-Tracqui, C; Crubézy, E; Ludes, B (August 2003). "Nuclear and Mitochondrial DNA Analysis of a 2,000-Year-Old Necropolis in the Egyin Gol Valley of Mongolia". Am. J. Hum. Genet. 73 (2): 247–60. doi:10.1086/377005. PMID 12858290.[|permanent dead link|dead link}}]
- ↑ 18.0 18.1 18.2 ИЗУЧЕНИЕ ГЕНЕТИЧЕСКОЙ СТРУКТУРЫ НАРОДОВ ЗАПАДНОГО КАВКАЗА ПО ДАННЫМ О ПОЛИМОРФИЗМЕ Y-ХРОМОСОМЫ, МИТОХОНДРИАЛЬНОЙ ДНК И ALU-ИНСЕРЦИЙ
- ↑ 19.0 19.1 19.2 Nasidze Ivan; Ling, E. Y. S.; Quinque, D.; Dupanloup, I.; Cordaux, R.; Rychkov, S.; Naumova, O.; Zhukova, O. et al. (2004). "Mitochondrial DNA and Y-Chromosome Variation in the Caucasus". Annals of Human Genetics 68 (Pt 3): 205–221. doi:10.1046/j.1529-8817.2004.00092.x. PMID 15180701. https://www.familytreedna.com/pdf/caucasus.pdf. Retrieved 2016-10-09.
- ↑ Yunusbayev 2011
- ↑ Haber, Marc; Platt, Daniel E.; Bonab, Maziar Ashrafian; Youhanna, Sonia C.; Soria-Hernanz, David F.; Martínez-Cruz, Begoña; Douaihy, Bouchra; Ghassibe-Sabbagh, Michella et al. (2012). "Afghanistan's Ethnic Groups Share a Y-Chromosomal Heritage Structured by Historical Events". PLOS ONE 7 (3): e34288. doi:10.1371/journal.pone.0034288. ISSN 1932-6203. PMID 22470552. Bibcode: 2012PLoSO...734288H.
- ↑ Haber, MExpression error: Unrecognized word "etal". (2012). "Afghanistan's Ethnic Groups Share a Y-Chromosomal Heritage Structured by Historical Events". PLOS ONE 7 (3): e34288. doi:10.1371/journal.pone.0034288. PMID 22470552. Bibcode: 2012PLoSO...734288H.
- ↑ 23.0 23.1 23.2 23.3 Afghan Hindu Kush: Where Eurasian Sub-Continent Gene Flows Converge
- ↑ 24.0 24.1 24.2 24.3 24.4 Introducing the Algerian Mitochondrial DNA and Y-Chromosome Profiles into the North African Landscape
- ↑ "Armenian DNA Project". http://f6.s.qip.ru/SeF39xoH.jpg.
- ↑ 26.0 26.1 26.2 "Where did European men come from?". 2008. http://www.jogg.info/41/Wiik.pdf.
- ↑ "Comments on 2014 Klyosov Article on Jewish DNA Genealogy p. 1 of 2 - Levite DNA". https://sites.google.com/site/levitedna/origins-of-r1a1a-ashkenazi-levites/2014-klyosov-article-on-jewish-dna-genealogy/comments-on-2014-klyosovarticle-on-jewish-dna-genealogy.
- ↑ 28.0 28.1 Кутуев Ильдус Альбертович. Генетическая структура и молекулярная филогеография народов кавказа
- ↑ ВКЛАД ОТДЕЛЬНЫХ ПОЛЕССКИХ ПОПУЛЯЦИЙ И ПОПУЛЯЦИИ БЕЛОРУССКИХ ТАТАР В ГЕНОФОНД НАСЕЛЕНИЯ БЕЛАРУСИ [1]
- ↑ 30.0 30.1 30.2 "High-resolution phylogenetic analysis of southeastern Europe traces major episodes of paternal gene flow among Slavic populations". Mol. Biol. Evol. 22 (10): 1964–75. October 2005. doi:10.1093/molbev/msi185. PMID 15944443.
- ↑ 31.0 31.1 Battaglia, Vincenza; Fornarino, Simona; Al-Zahery, Nadia; Olivieri, Anna; Pala, Maria; Myres, Natalie M; King, Roy J; Rootsi, Siiri et al. (24 December 2008). "Y-chromosomal evidence of the cultural diffusion of agriculture in southeast Europe". European Journal of Human Genetics 17 (6): 820–30. doi:10.1038/ejhg.2008.249. PMID 19107149. PMC 2947100. http://www.draganprimorac.com/wp-content/uploads/2010/05/Battaglia.pdf.
- ↑ Karachanak, Sena; Fornarino, Simona; Grugni, Viola; Semino, Ornella; Toncheva, Draga; Galabov, Angel; Atanasov, Boris (2009). "Y-Chromosomal Haplogroups in Bulgarians". Comptes rendus de l'Académie bulgare des Sciences 62 (3): 393–400. INIST:21359873. ISSN 1310-1331. http://www.proceedings.bas.bg/cgi-bin/mitko/0DOC_abs.pl?2009_3_16.
- ↑ Pereira, Luisa Maria Sousa Mesquita, ed (2013). "Y-chromosome diversity in modern Bulgarians: new clues about their ancestry". PLOS ONE 8 (3): e56779. doi:10.1371/journal.pone.0056779. PMID 23483890. Bibcode: 2013PLoSO...856779K.
- ↑ "Bulgarian_Y_Table.xlsx". https://docs.google.com/spreadsheets/d/1-TYdFLe_4wYW7HrJEQo7lryJM4ag_m4nQ7tTLzzBIlQ/pubhtml.
- ↑ D. Primorac (2022). "Croatian genetic heritage: an updated Y-chromosome story". Croatian Medical Journal 63 (3): 273–286. doi:10.3325/cmj.2022.63.273. PMID 35722696. PMC 9284021. http://www.cmj.hr/2022/63/3/35722696.htm.
- ↑ 36.00 36.01 36.02 36.03 36.04 36.05 36.06 36.07 36.08 36.09 36.10 36.11 36.12 Geographical Structure of the Y-chromosomal Genetic Landscape of the Levant: A coastal-inland contrast
- ↑ Y-chromosomal variation in the Czech Republic
- ↑ Altena, E., Smeding, R., van der Gaag, K.J. et al. The Dutch Y-chromosomal landscape. Eur J Hum Genet 28, 287–299 (2020). https://doi.org/10.1038/s41431-019-0496-0
- ↑ ZONEN VAN ADAM IN NEDERLAND Genetische genealogie : een zoektocht in ons DNA-archief
- ↑ El-Sibai, Mirvat; Platt, Daniel E.; Haber, Marc; Xue, Yali; Youhanna, Sonia C.; Wells, R. Spencer; Izaabel, Hassan; Sanyoura, May F. et al. (2009). "Geographical Structure of the Y-chromosomal Genetic Landscape of the Levant: A coastal-inland contrast". Annals of Human Genetics 73 (6): 568–581. doi:10.1111/j.1469-1809.2009.00538.x. ISSN 1469-1809. PMID 19686289.
- ↑ "Photo" (PNG). https://s-media-cache-ak0.pinimg.com/originals/d1/09/43/d109434a9e66aad55a8b10c4677dbba3.png.
- ↑ Migration Eaves to the Baltic Region
- ↑ "Y Haplogroup Frequencies in the Flemish Populstion". http://www.jogg.info/32/mertens.htm.
- ↑ Lappalainen, Tuuli; Koivumäki, Satu; Salmela, Elina; Huoponen, Kirsi; Sistonen, Pertti; Savontaus, Marja-Liisa; Lahermo, Päivi (19 July 2006). "Regional differences among the Finns: A Y-chromosomal perspective". Gene 376 (2): 207–215. doi:10.1016/j.gene.2006.03.004. PMID 16644145.
- ↑ Ramos-Luis, E.; Blanco-Verea, A.; Brión, M.; Van Huffel, V.; Carracedo, A.; Sánchez-Diz, P. (December 2009). "Phylogeography of French male lineages". Forensic Science International: Genetics Supplement Series 2 (1): 439–441. doi:10.1016/j.fsigss.2009.09.026.
- ↑ Kayser, Manfred; Lao, Oscar; Anslinger, K; Augustin, Christa; Bargel, G.; Edelmann, J; Elias, Sahar; Heinrich, M et al. (2005). "Significant genetic differentiation between Poland and Germany follows present-day political borders, as revealed by Y-chromosome analysis". Human Genetics 117 (5): 428–443. doi:10.1007/s00439-005-1333-9. ISSN 0340-6717. PMID 15959808. https://www.researchgate.net/publication/40852497.
- ↑ "Archived copy". https://www.familytreedna.com/pdf/italy.pdf.(Subscription content?)
- ↑ Sanchez, J.J.; Børsting, C.; Hernandez, A.; Mengel-Jørgensen, J.; Morling, N. (April 2004). "Y chromosome SNP haplogroups in Danes,Greenlanders and Somalis". International Congress Series 1261: 347–349. doi:10.1016/S0531-5131(03)01635-2.
- ↑ Presence of three different paternal lineages among North Indians: a study of 560 Y chromosomes (2009)
- ↑ 50.0 50.1 Influences of history, geography, and religion on genetic structure: the Maronites in Lebanon
- ↑ Tambets, KExpression error: Unrecognized word "etal". (April 2004). "The western and eastern roots of the Saami--the story of genetic "outliers" told by mitochondrial DNA and Y chromosomes". Am. J. Hum. Genet. 74 (4): 661–82. doi:10.1086/383203. PMID 15024688.
- ↑ A Y Chromosome Census of the British Isles
- ↑ Uniparental Markers in Italy Reveal a Sex-Biased Genetic Structure and Different Historical Strata
- ↑ Clinal patterns of human Y chromosomal diversity in continental Italy and Greece are dominated by drift and founder effects
- ↑ Traces of forgotten historical events in mountain communities in Central Italy: A genetic insight
- ↑ Uniparental Markers of Contemporary Italian Population Reveals Details on Its Pre-Roman Heritage
- ↑ Genetic Structure in Contemporary South Tyrolean Isolated Populations Revealed by Analysis of Y-Chromosome, mtDNA, and Alu Polymorphisms
- ↑ Differential Greek and northern African migrations to Sicily are supported by genetic evidence from the Y chromosome
- ↑ Isolates in a corridor of migrations: A high-resolution analysis of Y-chromosome variation in Jordan
- ↑ Вестник Московского университета. Серия XXIII АНТРОПОЛОГИЯ № 1/2014: 96–101СВЯЗЬ ИЗМЕНЧИВОСТИ Y ХРОМОСОМЫ И РОДОВОЙСТРУКТУРЫ: ГЕНОФОНД СТЕПНОЙ АРИСТОКРАТИИИ ДУХОВЕНСТВА КАЗАХОВ
- ↑ 61.0 61.1 Y chromosomes in Iranians and Tajiks
- ↑ 62.0 62.1 MtDNA and Y-chromosome Variation in Kurdish Groups
- ↑ The dual origin of tati-speakers from dagestan as written in the genealogy of uniparental variants
- ↑ Pliss et al. Y-Chromosomal Lineages of Latvians in the Contextof the Genetic Variation of the Eastern-Baltic Region
- ↑ Y-Chromosomal Diversity in Lebanon Is Structured by Recent Historical Events
- ↑ Paternal lineages in Libya inferred from Y-chromosome haplogroups
- ↑ Y-chromosomal evidence for a limited Greek contribution to the Pathan population of Pakistan (2006)
- ↑ Micro-Phylogeographic and Demographic History of Portuguese Male Lineages
- ↑ Kivisild, Toomas, ed (2012). "Y-chromosome analysis in individuals bearing the Basarab name of the first dynasty of Wallachian kings". PLOS ONE 7 (7): e41803. doi:10.1371/journal.pone.0041803. PMID 22848614. Bibcode: 2012PLoSO...741803M.
- ↑ Saudi Arabian Y-Chromosome diversity and its relationship with nearby regions.
- ↑ "Genetic characterization of 27 Y-STR loci with the Yfiler® Plus kit in the population of Serbia". Forensic Science International. Genetics 31: e48–e49. November 2017. doi:10.1016/j.fsigen.2017.07.013. PMID 28789900.
- ↑ Mihajlovic, Milica; Tanasic, Vanja; Markovic, Milica Keckarevic; Kecmanovic, Miljana; Keckarevic, Dusan (2022-11-01). "Distribution of Y-chromosome haplogroups in Serbian population groups originating from historically and geographically significant distinct parts of the Balkan Peninsula" (in English). Forensic Science International: Genetics 61: 102767. doi:10.1016/j.fsigen.2022.102767. ISSN 1872-4973. PMID 36037736. https://www.fsigenetics.com/article/S1872-4973(22)00108-9/abstract.
- ↑ The genetic structure of the Slovak population revealed by Y-chromosome polymorphisms
- ↑ ANALIZA Y-DNK HAPLOTIPOV SLOVENCEV
- ↑ Adams et al. The Genetic Legacy of Religious Diversity and Intolerance: Paternal Lineages of Christians, Jews, and Muslims in the Iberian Peninsula
- ↑ Y-Chromosome Variation Among Sudanese: Restricted Gene Flow, Concordance With Language, Geography, and History
- ↑ Karlsson, Andreas O; Wallerstrom, Thomas; Gotherstrom, Anders; Holmlund, Gunilla (2006). "Y-chromosome diversity in Sweden – A long-time perspective". European Journal of Human Genetics 14 (8): 963–970. doi:10.1038/sj.ejhg.5201651. PMID 16724001.
- ↑ "Phylogeography of Y-chromosome haplogroup I-M170 reveals distinct domains of prehistoric gene flow in europe". Am. J. Hum. Genet. 75 (1): 128–37. July 2004. doi:10.1086/422196. PMID 15162323.
- ↑ "Swedish Haplogroup Database (Stats haplopie)". http://dna.scangen.se/index.php?show=stats&stat=haplopie&lang=en&haplo_level=3&lan_sel=&database=ftdna.
- ↑ Y-chromosome distributions among populations in Northwest China identify significant contribution from Central Asian pastoralists and lesser influence of western Eurasians (2010)
- ↑ Bosch, E.; Calafell, F.; Gonzalez-Neira, A.; Flaiz, C; Mateu, E; Scheil, HG; Huckenbeck, W; Efremovska, L et al. (2006). "Paternal and maternal lineages in the Balkans show a homogeneous landscape over linguistic barriers, except for the isolated Aromuns". Annals of Human Genetics 70 (Pt 4): 459–87. doi:10.1111/j.1469-1809.2005.00251.x. PMID 16759179.
- ↑ Lappalainen, T.; Laitinen, V.; Salmela, E.; Andersen, P.; Huoponen, K.; Savontaus, M.-L.; Lahermo, P. (May 2008). "Migration waves to the Baltic Sea region". Ann. Hum. Genet. 72 (Pt 3): 337–48. doi:10.1111/j.1469-1809.2007.00429.x. PMID 18294359.
- ↑ ISOGG 2011
- ↑ Cinnioğlu, Cengiz; King, Roy; Kivisild, Toomas; Kalfoğlu, Ersi; Atasoy, Sevil; Cavalleri, Gianpiero L.; Lillie, Anita S.; Roseman, Charles C. et al. (January 2004). "Excavating Y-chromosome haplotype strata in Anatolia" (in en). Human Genetics 114 (2): 127–148. doi:10.1007/s00439-003-1031-4. ISSN 0340-6717. PMID 14586639.
- ↑ "ISOGG 2017 Y-DNA Haplogroup I". http://isogg.org/tree/ISOGG_HapgrpI.html.
- ↑ 86.0 86.1 Caciagli, Laura; Bulayeva, Kazima; Bulayev, Oleg; Bertoncini, Stefania; Taglioli, Luca; Pagani, Luca; Paoli, Giorgio; Tofanelli, Sergio (2009). "The key role of patrilineal inheritance in shaping the genetic variation of Dagestan highlanders" (in en). Journal of Human Genetics 54 (12): 689–694. doi:10.1038/jhg.2009.94. ISSN 1434-5161. PMID 19911015.
- ↑ Rootsi, Siiri; Magri, Chiara; Kivisild, Toomas; Benuzzi, Giorgia; Help, Hela; Bermisheva, Marina; Kutuev, Ildus; Barać, Lovorka et al. (2004). "Phylogeography of Y-Chromosome Haplogroup I Reveals Distinct Domains of Prehistoric Gene Flow in Europe". American Journal of Human Genetics 75 (1): 128–137. doi:10.1086/422196. ISSN 0002-9297. PMID 15162323.
- ↑ "Afghan Hindu Kush: Where Eurasian Sub-Continent Gene Flows Converge". PLOS ONE 8 (10): e76748. October 2013. doi:10.1371/journal.pone.0076748. PMID 24204668. Bibcode: 2013PLoSO...876748D.
- ↑ Allentoft, Morten E.; Sikora, Martin; Sjögren, Karl-Göran; Rasmussen, Simon; Rasmussen, Morten; Stenderup, Jesper; Damgaard, Peter B.; Schroeder, Hannes et al. (June 2015). "Population genomics of Bronze Age Eurasia" (in en). Nature 522 (7555): 167–172. doi:10.1038/nature14507. ISSN 1476-4687. PMID 26062507. Bibcode: 2015Natur.522..167A. https://www.nature.com/articles/nature14507.
- ↑ Malmström, Helena; Gilbert, M. Thomas P.; Thomas, Mark G.; Brandström, Mikael; Storå, Jan; Molnar, Petra; Andersen, Pernille K.; Bendixen, Christian et al. (2009-11-03). "Ancient DNA Reveals Lack of Continuity between Neolithic Hunter-Gatherers and Contemporary Scandinavians" (in en). Current Biology 19 (20): 1758–1762. doi:10.1016/j.cub.2009.09.017. ISSN 0960-9822. PMID 19781941.
- ↑ Karlsson, Andreas O.; Wallerström, Thomas; Götherström, Anders; Holmlund, Gunilla (August 2006). "Y-chromosome diversity in Sweden – A long-time perspective" (in en). European Journal of Human Genetics 14 (8): 963–970. doi:10.1038/sj.ejhg.5201651. ISSN 1476-5438. PMID 16724001.
- ↑ 92.0 92.1 92.2 Rootsi. "Phylogeography of Y-Chromosome Haplogroup I Reveals Distinct Domains of Prehistoric Gene Flow in Europe figure 1". http://hpgl.stanford.edu/publications/AJHG_2004_v75_Semino.pdf.
- ↑ "The peopling of modern Bosnia-Herzegovina: Y-chromosome haplogroups in the three main ethnic groups". Ann. Hum. Genet. 69 (Pt 6): 757–63. November 2005. doi:10.1111/j.1529-8817.2005.00190.x. PMID 16266413. http://www3.interscience.wiley.com/resolve/openurl?genre=article&sid=nlm:pubmed&issn=0003-4800&date=2005&volume=69&issue=Pt%206&spage=757.[|permanent dead link|dead link}}]
- ↑ O.M. Utevska (2017). Генофонд українців за різними системами генетичних маркерів: походження і місце на європейському генетичному просторі [The gene pool of Ukrainians revealed by different systems of genetic markers: the origin and statement in Europe] (PhD) (in українська). National Research Center for Radiation Medicine of National Academy of Sciences of Ukraine. pp. 219–226, 302.
- ↑ McEvoy and Bradley, Brian P and Daniel G (2010). Celtic from the West Chapter 5: Irish Genetics and Celts. Oxbow Books, Oxford, UK. p. 117. ISBN 978-1-84217-410-4.
- ↑ Haber, Marc; Platt, Daniel E.; Ashrafian Bonab, Maziar; Youhanna, Sonia C.; Soria-Hernanz, David F.; Martínez-Cruz, Begoña; Douaihy, Bouchra; Ghassibe-Sabbagh, Michella et al. (2012). "Afghanistan's ethnic groups share a Y-chromosomal heritage structured by historical events". PLOS ONE 7 (3): e34288. doi:10.1371/journal.pone.0034288. PMID 22470552. Bibcode: 2012PLoSO...734288H.
- ↑ Grasgruber, Pavel; Popović, Stevo; Bokuvka, Dominik; Davidović, Ivan; Hřebíčková, Sylva; Ingrová, Pavlína; Potpara, Predrag; Prce, Stipan et al. (2017). "The mountains of giants: An anthropometric survey of male youths in Bosnia and Herzegovina" (in en). Royal Society Open Science 4 (4): 161054. doi:10.1098/rsos.161054. PMID 28484621. Bibcode: 2017RSOS....461054G.
- ↑ Grasgruber, P.; Cacek, J.; Kalina, T.; Sebera, M. (2014-12-01). "The role of nutrition and genetics as key determinants of the positive height trend" (in en). Economics & Human Biology 15: 81–100. doi:10.1016/j.ehb.2014.07.002. ISSN 1570-677X. PMID 25190282.
External links
Phylogenetic tree and distribution maps
- Y-DNA Haplogroup I-M170 and Its Subclades from the International Society of Genetic Genealogy (ISOGG)of 2013
- Phylogeography of Y-Chromosome Haplogroup I
- Frequency Distributions of Y-DNA Haplogroup I and its subclades – with Video Tutorial
- Frequency and Variance of I1b (now considered I2a2-M26)
- Map of 'I1a' (now considered I-M253)
- Map of 'I1b' (now considered I2a-P37.2)
- Map of 'I1c' (now considered I2b-M223)
- Rescalled Haplogroup I Tree (K. Nordtvedt 2011).
Projects
- I Project at FTDNA
- I1 Project at FTDNA
- I2* Project at FTDNA
- I2a project at FTDNA
- I2b project at FTDNA
- I2b2 L38+ project at FTDNA
- The Scandinavian yDNA Genealogical Project at FTDNA
- The Finland Genealogical Project at FTDNA
Other
- Study of Y-Haplogroup I and Modal Haplotypes
- The Y Chromosome Consortium (YCC)
- Example haplotypes from I1* "y cluster"
- YCC Haplogroup I page – I1a (now considered I-M253), I1b (now considered I-P37.2) and I1c (now considered I-M223)
- Haplo-I Subclade Predictor
- Spread of Haplogroup I, from National Geographic
- I2b2 Y-DNA found in Bronze Age skeletons of Lichtenstein Cave
- Haplogroup I-L38 (I2b2) In Search of the Origin of I-L38 (aka I2b2)
 |