Biology:Haplogroup J (Y-DNA)
| Haplogroup J-M304 | |
|---|---|
 | |
| Possible time of origin | 42,900 years ago[1] |
| Coalescence age | 31,600 years ago[1] |
| Possible place of origin | Western Asia, Caucasus |
| Ancestor | IJ |
| Descendants | J-M172, J-M267 |
| Defining mutations | M304/Page16/PF4609, 12f2.1 |
| Highest frequencies | Ingush, Chechens, Avars, Dargins, Arabs, Assyrians, Jews, Greeks, Georgians, Arameans, Melkites, Mandeans, Italians, Cypriots |
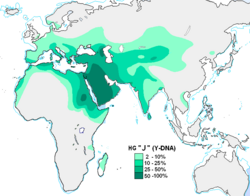
Haplogroup J-M304, also known as J,[Phylogenetics 1] is a human Y-chromosome DNA haplogroup. It is believed to have evolved in Western Asia.[2] The clade spread from there during the Neolithic, primarily into North Africa, the Horn of Africa, the Socotra Archipelago, the Caucasus, Europe, Anatolia, Central Asia, South Asia, and Southeast Asia.
Haplogroup J-M304 is divided into two main subclades (branches), J-M267 and J-M172.
Origins
Haplogroup J-M304 is believed to have split from the haplogroup I-M170 roughly 43,000 years ago in Western Asia,[1] as both lineages are haplogroup IJ subclades. Haplogroup IJ and haplogroup K derive from haplogroup IJK, and only at this level of classification does haplogroup IJK join with Haplogroup G-M201 and Haplogroup H as immediate descendants of Haplogroup F-M89. J-M304 (Transcaucasian origin) is defined by the M304 genetic marker, or the equivalent 12f2.1 marker. The main current subgroups J-M267 (Armenia highlands origin) and J-M172 (Zagros mountains origin), which now comprise between them almost all of the haplogroup's descendant lineages, are both believed to have arisen very early, at least 10,000 years ago. Nonetheless, Y-chromosomes F-M89* and IJ-M429* were reported to have been observed in the Iranian plateau (Grugni et al. 2012).
On the other hand, it would seem to be that different episodes of populace movement had impacted southeast Europe, as well as the role of the Balkans as a long-standing corridor to Europe from the Near East is shown by the phylogenetic unification of Hgs I and J by the basal M429 mutation. This proof of common ancestry suggests that ancestral Hgs IJ-M429* probably would have entered Europe through the Balkan track sometime before the LGM. They then subsequently split into Hg J and Hg I in Middle East and Europe in a typical disjunctive phylogeographic pattern. Such a geographic hall[clarification needed] is prone to have encountered extra consequent gene streams, including the horticultural settlers. Moreover, the unification of haplogroups IJK creates evolutionary distance from F–H delegates, as well as supporting the inference that both IJ-M429 and KT-M9 arose closer to the Middle East than Central or East Asia.[citation needed]
Haplogroup J has also been found among two ancient Egyptian mummies excavated at the Abusir el-Meleq archaeological site in Middle Egypt, which date from a period between the late New Kingdom and the Roman era.[3]
Distribution
Haplogroup J-M267 is found in its greatest concentration in the Arabian peninsula. Outside of this region, haplogroup J-M304 has a significant presence in other parts of the Middle East as well as in North Africa, the Horn of Africa, and Caucasus. It also has a moderate occurrence in Southern Europe, especially in central and southern Italy, Malta, Greece and Albania. The J-M410 subclade is mostly distributed in Anatolia, Greece and southern Italy. Additionally, J-M304 is observed in Central Asia and South Asia, particularly in the form of its subclade J-M172. J-12f2 and J-P19 are also found among the Herero (8%).[4]
| Country/Region | Sampling | N | J-M267 | J-M172 | Total J | Study |
|---|---|---|---|---|---|---|
| Algeria | Oran | 102 | 22.5 | 4.9 | 27.4 | Robino 2008 |
| Albania | Tirana | 30 | 20.0 | Bosch 2006 | ||
| Albania | 55 | 23.64 | Battaglia 2008 | |||
| Bosnia | Serbs | 81 | 9.9 | Battaglia 2008 | ||
| Caucasus | Chechen | 330 | 20.9 | 56.7 | 77.6 | Balanovsky 2011 |
| Caucasus | Ingush | 143 | 2.8 | 88.8 | 91.6 | Balanovsky 2011 |
| China | Uygur | 50 | 0 | 34.0 | 34.0 | Shou 2010 |
| China | Uzbek | 23 | 0 | 30.4 | 34.7 | Shou 2010 |
| China | Tajik | 31 | 0 | 16.1 | 16.1 | Shou 2010 |
| China | Han Chinese | 30 | 10 | 10 | Xue 2006 | |
| Cyprus | 164 | 9.6 | 12.9 | 22.5 | El-Sibai 2009[5] | |
| Egypt | 124 | 19.8 | 7.6 | 27.4 | El-Sibai 2009 | |
| Greece | Crete/Heraklion | 104 | 1.9 | 44.2 | 46.1 | Martinez 2007 |
| Greece | Crete | 143 | 3.5 | 35 | 38.5 | El-Sibai 2009 |
| Greece | 154 | 1.9 | 18.1 | 20 | El-Sibai 2009 | |
| India | Sunni and North Indian Shia | 112 | 32 | 43.2 | 75.2 | El-Sibai 2009 |
| Iran | 92 | 3.2 | 25 | 28.2 | El-Sibai 2009 | |
| Iraq | Arab and Assyrian | 117 | 33.1 | 25.1 | 58.2 | El-Sibai 2009 |
| Israel | Akko (Arabs) | 101 | 39.2 | 18.6 | 57.8 | El-Sibai 2009 |
| Italy | 699 | 2 | 20 | 22 | Capelli 2007 | |
| Italy | Central Marche | 59 | 5.1 | 35.6 | 40.7 | Capelli 2007 |
| Italy | West Calabria | 57 | 3.5 | 35.1 | 38.6 | Capelli 2007 |
| Italy | Sicily | 212 | 5.2 | 22.6 | 27.8 | El-Sibai 2009 |
| Italy | Sardinia | 81 | 4.9 | 9.9 | 14.8 | El-Sibai 2009 |
| Jordan | 273 | 35.5 | 14.6 | 50.1 | El-Sibai 2009 | |
| Kosovo | Albanians | 114 | 16.67 | Pericić 2005 | ||
| Kuwait | 42 | 33.3 | 9.5 | 42.8 | El-Sibai 2009 | |
| Lebanon | 951 | 17 | 29.4 | 46.4 | El-Sibai 2009 | |
| Malta | 90 | 7.8 | 21.1 | 28.9 | El-Sibai 2009 | |
| Morocco | 316 | 1 | 0.2 | 1.2 | El-Sibai 2009 | |
| Morocco | Residents in Italy | 51 | 19.6 | 0 | 19.6 | Onofri 2008 |
| Portugal | Portugal | 303 | 4.3 | 6.9 | 11.2 | El-Sibai 2009 |
| Qatar | Qatar | 72 | 58.3 | 8.3 | 66.6 | El-Sibai 2009 |
| Saudi Arabia | 157 | 40.13 | 15.92 | 57.96 | Abu-Amero 2009 | |
| Serbia | Belgrade | 113 | 8 | Pericić 2005 | ||
| Serbia | 179 | 5.6 | Mirabal 2010 | |||
| Spain | Cadiz | 28 | 3.6 | 14.3 | 17.9 | El-Sibai 2009 |
| Spain | Cantabria | 70 | 2.9 | 2.9 | 5.8 | El-Sibai 2009 |
| Spain | Castille | 21 | 0 | 9.5 | 9.5 | El-Sibai 2009 |
| Spain | Cordoba | 27 | 0 | 14.7 | 14.7 | El-Sibai 2009 |
| Spain | Galicia | 19 | 5.3 | 0 | 5.3 | El-Sibai 2009 |
| Spain | Huelva | 22 | 0 | 13.7 | 13.7 | El-Sibai 2009 |
| Spain | Ibiza | 54 | 0 | 3.7 | 3.7 | El-Sibai 2009 |
| Spain | Leon | 60 | 1.7 | 5 | 6.7 | El-Sibai 2009 |
| Spain | Malaga | 26 | 0 | 15.4 | 15.4 | El-Sibai 2009 |
| Spain | Mallorca | 62 | 1.6 | 8 | 9.7 | El-Sibai 2009 |
| Spain | Sevilla | 155 | 3.2 | 7.8 | 11 | El-Sibai 2009 |
| Spain | Valencia | 31 | 2.7 | 5.5 | 8.2 | El-Sibai 2009 |
| Syria | Arab and Assyrian | 554 | 33.6 | 20.8 | 54.4 | El-Sibai 2009 |
| Tunisia | 62 | 0 | 8 | 8 | El-Sibai 2009 | |
| Tunisia | 52 | 34.6 | 3.8 | 38.4 | Onofri 2008 | |
| Tunisia | Sousse | 220 | 25.9 | 8.2 | 34.1 | Fadhlaoui-Zid 2014 |
| Tunisia | Tunis | 148 | 32.4 | 3.4 | 35.8 | Arredi 2004 |
| Turkey | 523 | 9.1 | 24.2 | 33.3 | El-Sibai 2009 | |
| UAE | 164 | 34.7 | 10.3 | 45 | El-Sibai 2009 | |
| Yemen | 62 | 72.5 | 9.6 | 82.1 | El-Sibai 2009 |
Subclade distribution
J-M304*
Paragroup J-M304*[Phylogenetics 2] includes all of J-M304 except for J-M267, J-M172 and their subclades. J-M304* is rarely found outside of the island of Socotra, belonging to Yemen, where it is extremely frequent at 71.4% and j1-267 for the rest with no j2[6] Haplogroup J-M304* also has been found with lower frequency in Oman (Di Giacomo 2004), Ashkenazi Jews,[7] Saudi Arabia (Abu-Amero 2009), Greece (Di Giacomo 2004), the Czech Republic (Di Giacomo 2004 and Luca 2007), Uygurs[8] and several Turkic peoples.[9] (Cinnioglu 2004 and Varzari 2006).
YFull[1] and FTDNA[10] have however failed to find J* people anywhere in the world although there are 2 J2-Y130506 persons and 1 J1 person from Soqotra. But Cerny 2009 study found 9 J1 persons in Soqotra/Socotra and majority of J* and no J2, hypothesizing a J1 founder effect in Socotra.
The following gives a summary of most of the studies which specifically tested for J-M267 and J-M172, showing its distribution in Europe, North Africa, the Middle East and Central Asia.
J-M267
Haplogroup J-M267[Phylogenetics 3] defined by the M267 SNP is in modern times most frequent in the Arabian Peninsula: Yemen (up to 76%),[11] Saudi (up to 64%) (Alshamali 2009), Qatar (58%),[12] and Dagestan (up to 56%).[13] J-M267 is generally frequent among Arab Bedouins (62%),[14] Ashkenazi Jews (20%) (Semino 2004), Algeria (up to 35%) (Semino 2004), Iraq (28%) (Semino 2004), Tunisia (up to 31%),[15] Syria (up to 30%), Egypt (up to 20%) (Luis 2004), and the Sinai Peninsula. To some extent, the frequency of Haplogroup J-M267 collapses at the borders of Arabic/Semitic-speaking territories with mainly non-Arabic/Semitic speaking territories, such as Turkey (9%), Iran (5%), Sunni Indian Muslims (2.3%) and Northern Indian Shia (11%) (Eaaswarkhanth 2009). Some figures above tend to be the larger ones obtained in some studies, while the smaller figures obtained in other studies are omitted. It is also highly frequent among Jews, especially the Kohanim line (46%) (Hammer 2009).
ISOGG states that J-M267 originated in the Middle East. It is found in parts of the Near East, Anatolia and North Africa, with a much sparser distribution in the southern Mediterranean flank of Europe, and in Ethiopia.
But not all studies agree on the point of origin. The Levant has been proposed but a 2010 study concluded that the haplogroup had a more northern origin, possibly Anatolia.
The origin of the J-P58 subclade is likely in the more northerly populations and then spreads southward into the Arabian Peninsula. The high Y-STR variance of J-P58 in ethnic groups in Turkey, as well as northern regions in Syria and Iraq, supports the inference of an origin of J-P58 in nearby eastern Anatolia. Moreover, the network analysis of J-P58 haplotypes shows that some of the populations with low diversity, such as Bedouins from Israel, Qatar, Sudan and the United Arab Emirates, are tightly clustered near high-frequency haplotypes. This suggests that founder effects with star burst expansion into the Arabian Desert (Chiaroni 2010).
J-M172
Haplogroup J-M172[Phylogenetics 4] is found in the highest concentrations in the Caucasus and the Fertile Crescent/Iraq and is found throughout the Mediterranean (including the Italian, Balkan, Anatolian and Iberian peninsulas and North Africa) (Di Giacomo 2003).
The highest ever reported concentration of J-M172 was 72% in Northeastern Georgia (Nasidze 2004). Other high reports include Ingush 32% (Nasidze 2004), Cypriots 30-37% (Capelli 2005), Lebanese 30% (Wells et al. 2001), Assyrian, Mandean and Arab Iraqis 29.7% (Sanchez et al. 2005)[full citation needed], Syrians and Syriacs 22.5%, Kurds 24%-28%, Pashtuns 20-30%,[16]Iranians 23% (Aburto 2006), Ashkenazi Jews 24%, Palestinian Arabs 16.8%-25%, Sephardic Jews 29%[17] and North Indian Shia Muslim 18%, Chechens 26%, Balkars 24%, Yaghnobis 32%, Armenians 21-24%, and Azerbaijanis 24%-48%.
In South Asia, J2-M172 was found to be significantly higher among Dravidian castes at 19% than among Indo-European castes at 11%. J2-M172 and J-M410 is found 21% among Dravidian middle castes, followed by upper castes, 18.6%, and lower castes 14%. (Sengupta 2006)[18] Subclades of M172 such as M67 and M92 were not found in either Indian or Pakistani samples which also might hint at a partial common origin.(Sengupta 2006)[18]
According to a genetic study in China by Shou et al., J2-M172 is found with high frequency among Uygurs (17/50 = 34%) and Uzbeks (7/23 = 30.4%), moderate frequency among Pamiris (5/31 = 16.1%), and also found J-M172 in Han Chinese (10%)[19] and low frequency among Yugurs (2/32 = 6.3%) and Monguors (1/50 = 2.0%). The authors also found J-M304(xJ2-M172) with low frequency among the Russians (1/19 = 5.3%), Uzbeks (1/23 = 4.3%), Sibe people (1/32 = 3.1%), Dongxiangs (1/35 = 2.9%), and Kazakhs (1/41 = 2.4%) in Northwest China.[20] Only far northwestern ethnic minorities had haplogroup J in Xinjiang, China. Uzbeks in the sample had 30.4% J2-M172 and Tajiks of Xinjiang and Uyghurs also had it.[21]
Phylogenetics
In Y-chromosome phylogenetics, subclades are the branches of haplogroups. These subclades are also defined by single nucleotide polymorphisms (SNPs) or unique event polymorphisms (UEPs).
Phylogenetic history
Prior to 2002, there were in academic literature at least seven naming systems for the Y-Chromosome Phylogenetic tree. This led to considerable confusion. In 2002, the major research groups came together and formed the Y-Chromosome Consortium (YCC). They published a joint paper that created a single new tree that all agreed to use. Later, a group of citizen scientists with an interest in population genetics and genetic genealogy formed a working group to create an amateur tree aiming at being above all timely. The table below brings together all of these works at the point of the landmark 2002 YCC Tree. This allows a researcher reviewing older published literature to quickly move between nomenclatures.
| YCC 2002/2008 (Shorthand) | (α) | (β) | (γ) | (δ) | (ε) | (ζ) | (η) | YCC 2002 (Longhand) | YCC 2005 (Longhand) | YCC 2008 (Longhand) | YCC 2010r (Longhand) | ISOGG 2006 | ISOGG 2007 | ISOGG 2008 | ISOGG 2009 | ISOGG 2010 | ISOGG 2011 | ISOGG 2012 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| J-12f2a | 9 | VI | Med | 23 | Eu10 | H4 | B | J* | J | J | J | - | - | - | - | - | - | J |
| J-M62 | 9 | VI | Med | 23 | Eu10 | H4 | B | J1 | J1a | J1a | J1a | - | - | - | - | - | - | Private |
| J-M172 | 9 | VI | Med | 24 | Eu9 | H4 | B | J2* | J2 | J2 | J2 | - | - | - | - | - | - | J2 |
| J-M47 | 9 | VI | Med | 24 | Eu9 | H4 | B | J2a | J2a | J2a1 | J2a4a | - | - | - | - | - | - | J2a1a |
| J-M68 | 9 | VI | Med | 24 | Eu9 | H4 | B | J2b | J2b | J2a3 | J2a4c | - | - | - | - | - | - | J2a1c |
| J-M137 | 9 | VI | Med | 24 | Eu9 | H4 | B | J2c | J2c | J2a4 | J2a4h2a1 | - | - | - | - | - | - | J2a1h2a1a |
| J-M158 | 9 | VI | Med | 24 | Eu9 | H4 | B | J2d | J2d | J2a5 | J2a4h1 | - | - | - | - | - | - | J2a1h1 |
| J-M12 | 9 | VI | Med | 24 | Eu9 | H4 | B | J2e* | J2e | J2b | J2b | - | - | - | - | - | - | J2b |
| J-M102 | 9 | VI | Med | 24 | Eu9 | H4 | B | J2e1* | J2e1 | J2b | J2b | - | - | - | - | - | - | J2b |
| J-M99 | 9 | VI | Med | 24 | Eu9 | H4 | B | J2e1a | J2e1a | J2b2a | J2b2a | - | - | - | - | - | - | Private |
| J-M67 | 9 | VI | Med | 24 | Eu9 | H4 | B | J2f* | J2f | J2a2 | J2a4b | - | - | - | - | - | - | J2a1b |
| J-M92 | 9 | VI | Med | 24 | Eu9 | H4 | B | J2f1 | J2f1 | J2a2a | J2a4b1 | - | - | - | - | - | - | J2a1b1 |
| J-M163 | 9 | VI | Med | 24 | Eu9 | H4 | B | J2f2 | J2f2 | J2a2b | J2a4b2 | - | - | - | - | - | - | Private |
Research publications
The following research teams per their publications were represented in the creation of the YCC tree.
Phylogenetic trees
There are several confirmed and proposed phylogenetic trees available for haplogroup J-M304. The scientifically accepted one is the Y-Chromosome Consortium (YCC) one published in Karafet 2008 and subsequently updated. A draft tree that shows emerging science is provided by Thomas Krahn at the Genomic Research Center in Houston, Texas. The International Society of Genetic Genealogy (ISOGG) also provides an amateur tree.
The Genomic Research Center draft tree
This is Thomas Krahn at the Genomic Research Center's Draft tree Proposed Tree for haplogroup J-P209 (Krahn FTDNA). For brevity, only the first three levels of subclades are shown.
- J-M304 12f2a, 12f2.1, M304, P209, L60, L134
- M267, L255, L321, L765, L814, L827, L1030
- M62
- M365.1
- L136, L572, L620
- M390
- P56
- P58, L815, L828
- L256
- Z1828, Z1829, Z1832, Z1833, Z1834, Z1836, Z1839, Z1840, Z1841, Z1843, Z1844
- Z1842
- L972
- M172, L228
- M410, L152, L212, L505, L532, L559
- M289
- L26, L27, L927
- L581
- M12, M102, M221, M314, L282
- M205
- M241
- M410, L152, L212, L505, L532, L559
- M267, L255, L321, L765, L814, L827, L1030
The Y-Chromosome Consortium tree
This is the official scientific tree produced by the Y-Chromosome Consortium (YCC). The last major update was in 2008 (Karafet 2008). Subsequent updates have been quarterly and biannual. The current (2022) version is of the 2019/2020 update.
Prominent members of J
- Ben Affleck[22]
- Qajar dynasty[23]
- Fifth dynasty of Egypt[24]
- Khabib Nurmagomedov[22]
- House of Saud[23]
- Vincent van Gogh[25][23]
- Rothschild family[22]
- Dustin Hoffman[23]
- Bernard Montgomery[22]
- John Stamos[22]
- Bernie Sanders[22]
- Dzhokhar Dudayev[22]
- Adam Sandler[22]
See also
Genetics
Y-DNA J subclades
References
- ↑ 1.0 1.1 1.2 1.3 "J YTree". https://yfull.com/tree/J/.
- ↑ Y-DNA Haplogroup J , ISOGG, 2015
- ↑ Schuenemann, Verena J. (2017). "Ancient Egyptian mummy genomes suggest an increase of Sub-Saharan African ancestry in post-Roman periods". Nature Communications 8: 15694. doi:10.1038/ncomms15694. PMID 28556824. Bibcode: 2017NatCo...815694S.
- ↑ Wood, Elizabeth T. (2005). "Contrasting patterns of Y chromosome and mtDNA variation in Africa: evidence for sex-biased demographic processes". European Journal of Human Genetics 13 (7): 867–876. doi:10.1038/sj.ejhg.5201408. PMID 15856073. http://sites.lsa.umich.edu/bis/wp-content/uploads/sites/171/2014/10/Wood-et-al.-2005-EJHG.pdf. Retrieved 24 September 2016.
- ↑ El-Sibai 2009 reported results from several studies : Di Giacomo 2003, Al-Zahery 2003, Flores 2004, Cinnioglu 2004, Capelli 2005, Goncalves 2005, Zalloua 2008, Cadenas 2008
- ↑ Černý 2009: J-12f2(xM267, M172)(45/63) Černý, Viktor (2009). "Out of Arabia—the settlement of island Socotra as revealed by mitochondrial and Y chromosome genetic diversity". American Journal of Physical Anthropology 138 (4): 439–447. doi:10.1002/ajpa.20960. PMID 19012329. http://ychrom.invint.net/upload/iblock/f30/Cerny%202009%20Out%20of%20ArabiarusThe%20Settlement%20of%20Island%20Soqotra%20as%20Revealed%20by%20Mitochondrial%20and%20Y.pdf. Retrieved 12 June 2016.
- ↑ Shen 2004: Haplogroup J-M304(xM267, M172) in 1/20 Ashkenazi Jews.
- ↑ Zhong et al (2011), Mol Biol Evol January 1, 2011 vol. 28 no. 1 717-727 , See Table[yes|permanent dead link|dead link}}].
- ↑ Yunusbaev 2006:Stats are for combined Dagestan ethnic groups see the Dagestan article for details. Dargins (91%), Avars (67%), Chamalins (67%), Lezgins (58%), Tabassarans (49%), Andis (37%), Assyrians (29%), Bagvalins (21.4%))
- ↑ "FamilyTreeDNA - Y-DNA J Haplogroup Project". https://www.familytreedna.com/public/Y-DNA_J/default.aspx?section=yresults.
- ↑
- Alshamali 2009: 81% (84/104)
- Cadenas 2008: 45/62=72.6% J-M267
- ↑ Cadenas 2008: 42/72=58.3% J-M267
- ↑ Yunusbaev 2006: Dargwas (91%), Avars (67%), Chamalins (67%), Lezgins (58%), Tabassarans (49%), Andis (37%), Assyrians (29%), Bagvalins (21.4%))stats combined Dagestan ethnic groups see Dagestan article
- ↑ Nebel 2001: 21/32
- ↑ 31% is based on Combined Data
- Semino 2004: 30%
- Arredi 2004: 32%
- ↑ Haber, Marc; Platt, Daniel E.; Ashrafian Bonab, Maziar; Youhanna, Sonia C.; Soria-Hernanz, David F.; Martínez-Cruz, Begoña; Douaihy, Bouchra; Ghassibe-Sabbagh, Michella et al. (2012). "Afghanistan's Ethnic Groups Share a Y-Chromosomal Heritage Structured by Historical Events". PLOS ONE 7 (3): e34288. doi:10.1371/journal.pone.0034288. PMID 22470552. Bibcode: 2012PLoSO...734288H.
- ↑ "Volume 69". http://www.sciencedirect.com/science?_ob=MImg&_imagekey=B8JDD-4R29JBW-M-M&_cdi=43612&_user=1464331&_coverDate=11%2F30%2F2001&_sk=%23TOC%2343612%232001%23999309994%23673102%23FLA%23display%23Volume_69%2C_Issue_5%2C_Pages_i-ii%2C_923-1160_%28November_2001%29%23tagged%23Volume%23first%3D69%23Issue%23first%3D5%23date%23%28November_2001%29%23&view=c&_gw=y&wchp=dGLzVlz-zSkWA&md5=0d7ca8f01e76959310fd630f4ee8b621&ie=%2Fsdarticle.pdf.
- ↑ 18.0 18.1 Sengupta, SExpression error: Unrecognized word "etal". (February 2006). "Polarity and temporality of high-resolution y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists". Am. J. Hum. Genet. 78 (2): 202–21. doi:10.1086/499411. PMID 16400607.
- ↑ Xue 2006
- ↑ Shou et al (2010), Y-chromosome distributions among populations in Northwest China identify significant contribution from Central Asian pastoralists and lesser influence of western Eurasians , Journal of Human Genetics (2010) 55, 314–322; doi:10.1038/jhg.2010.30; published online 23 April 2010, Table 2. Haplogroup distribution and Y-chromosome diversity in 14 northwestern populations
- ↑ Shou, Wei-Hua; Qiao, Wn-Fa; Wei, Chuan-Yu; Dong, Yong-Li; Tan, Si-Jie; Shi, Hong; Tang, Wen-Ru; Xiao, Chun-Jie (2010). "Y-chromosome distributions among populations in Northwest China identify significant contribution from Central Asian pastoralists and lesser influence of western Eurasians". J Hum Genet 55 (5): 314–322. doi:10.1038/jhg.2010.30. PMID 20414255.
- ↑ 22.0 22.1 22.2 22.3 22.4 22.5 22.6 22.7 Maciamo. "Eupedia" (in en). http://www.eupedia.com/europe/Haplogroup_J2_Y-DNA.shtml.
- ↑ 23.0 23.1 23.2 23.3 Maciamo. "Eupedia" (in en). http://www.eupedia.com/europe/Haplogroup_J1_Y-DNA.shtml.
- ↑ Schuenemann, V. J.; Peltzer, A.; Welte, B.; Van Pelt, W. P.; Molak, M.; Wang, C. C.; Furtwängler, A.; Urban, C. et al. (2017). "Ancient Egyptian mummy genomes suggest an increase of Sub-Saharan African ancestry in post-Roman periods". Nature Communications 8: 15694. doi:10.1038/ncomms15694. PMID 28556824. Bibcode: 2017NatCo...815694S.
- ↑ "Welcome to FamilyTreeDNA Discover (Beta)" (in en). https://discover.familytreedna.com/.
Works cited
Journals
- Y Chromosome Consortium "YCC" (2002). "A Nomenclature System for the Tree of Human Y-Chromosomal Binary Haplogroups". Genome Research 12 (2): 339–48. doi:10.1101/gr.217602. PMID 11827954.
- Abu-Amero, Khaled K; Hellani, Ali; González, Ana M; Larruga, Jose M; Cabrera, Vicente M; Underhill, Peter A (2009). "Saudi Arabian Y-Chromosome diversity and its relationship with nearby regions". BMC Genetics 10: 59. doi:10.1186/1471-2156-10-59. PMID 19772609.
- Al-Zahery, N.; Semino, O.; Benuzzi, G.; Magri, C.; Passarino, G.; Torroni, A.; Santachiara-Benerecetti, A.S. (September 2003). "Y-chromosome and mtDNA polymorphisms in Iraq, a crossroad of the early human dispersal and of post-Neolithic migrations". Molecular Phylogenetics and Evolution 28 (3): 458–472. doi:10.1016/S1055-7903(03)00039-3. PMID 12927131.
- Alshamali, Farida; Pereira, Luísa; Budowle, Bruce; Poloni, Estella S.; Currat, Mathias (2009). "Local Population Structure in Arabian Peninsula Revealed by Y-STR diversity". Human Heredity 68 (1): 45–54. doi:10.1159/000210448. PMID 19339785.
- Arredi, B; Poloni, E; Paracchini, S; Zerjal, T; Fathallah, D; Makrelouf, M; Pascali, V; Novelletto, A et al. (2004). "A predominantly neolithic origin for Y-chromosomal DNA variation in North Africa". American Journal of Human Genetics 75 (2): 338–45. doi:10.1086/423147. PMID 15202071.
- Balanovsky, O.; Dibirova, K.; Dybo, A.; Mudrak, O.; Frolova, S.; Pocheshkhova, E.; Haber, M.; Platt, D. et al. (2011). "Parallel evolution of genes and languages in the Caucasus region". Molecular Biology and Evolution 28 (10): 2905–20. doi:10.1093/molbev/msr126. PMID 21571925.
- Battaglia, Vincenza; Fornarino, Simona; Al-Zahery, Nadia; Olivieri, Anna; Pala, Maria; Myres, Natalie M; King, Roy J; Rootsi, Siiri et al. (2009). "Y-chromosomal evidence of the cultural diffusion of agriculture in southeast Europe". European Journal of Human Genetics 17 (6): 820–30. doi:10.1038/ejhg.2008.249. PMID 19107149.
- Bosch, E.; Calafell, F.; Gonzalez-Neira, A.; Flaiz, C.; Mateu, E.; Scheil, H.-G.; Huckenbeck, W.; Efremovska, L. et al. (July 2006). "Paternal and maternal lineages in the Balkans show a homogeneous landscape over linguistic barriers, except for the isolated Aromuns". Annals of Human Genetics 70 (4): 459–487. doi:10.1111/j.1469-1809.2005.00251.x. PMID 16759179.
- Cadenas, Alicia M; Zhivotovsky, Lev A; Cavalli-Sforza, Luca L; Underhill, Peter A; Herrera, Rene J (2008). "Y-chromosome diversity characterizes the Gulf of Oman". European Journal of Human Genetics 16 (3): 374–86. doi:10.1038/sj.ejhg.5201934. PMID 17928816.
- Capelli, C.; Redhead, N.; Romano, V.; Cali, F.; Lefranc, G.; Delague, V.; Megarbane, A.; Felice, A. E. et al. (March 2006). "Population Structure in the Mediterranean Basin: A Y Chromosome Perspective". Annals of Human Genetics 70 (2): 207–225. doi:10.1111/j.1529-8817.2005.00224.x. PMID 16626331.
- Capelli, Cristian; Brisighelli, Francesca; Scarnicci, Francesca; Arredi, Barbara; Caglia’, Alessandra; Vetrugno, Giuseppe; Tofanelli, Sergio; Onofri, Valerio et al. (July 2007). "Y chromosome genetic variation in the Italian peninsula is clinal and supports an admixture model for the Mesolithic–Neolithic encounter". Molecular Phylogenetics and Evolution 44 (1): 228–239. doi:10.1016/j.ympev.2006.11.030. PMID 17275346.
- Chiaroni, Jacques; King, Roy J; Myres, Natalie M; Henn, Brenna M; Ducourneau, Axel; Mitchell, Michael J; Boetsch, Gilles; Sheikha, Issa et al. (2009). "The emergence of Y-chromosome haplogroup J1e among Arabic-speaking populations". European Journal of Human Genetics 18 (3): 348–53. doi:10.1038/ejhg.2009.166. PMID 19826455.
- Chiaroni, Jacques; King, Roy J; Myres, Natalie M; Henn, Brenna M; Ducourneau, Axel; Mitchell, Michael J; Boetsch, Gilles; Sheikha, Issa et al. (2010). "The emergence of Y-chromosome haplogroup J1e among Arabic-speaking populations". European Journal of Human Genetics 18 (3): 348–53. doi:10.1038/ejhg.2009.166. PMID 19826455.
- Cinnioglu, Cengiz; King, Roy; Kivisild, Toomas; Kalfoglu, Ersi; Atasoy, Sevil; Cavalleri, Gianpiero L.; Lillie, Anita S.; Roseman, Charles C. et al. (2004). "Excavating Y-chromosome haplotype strata in Anatolia". Human Genetics 114 (2): 127–48. doi:10.1007/s00439-003-1031-4. PMID 14586639.
- Di Giacomo, F.; Luca, F.; Anagnou, N.; Ciavarella, G.; Corbo, R.M.; Cresta, M.; Cucci, F.; Di Stasi, L. et al. (2003). "Clinal patterns of human Y chromosomal diversity in continental Italy and Greece are dominated by drift and founder effects". Molecular Phylogenetics and Evolution 28 (3): 387–95. doi:10.1016/S1055-7903(03)00016-2. PMID 12927125.
- Di Giacomo, F.; Luca, F.; Popa, L. O.; Akar, N.; Anagnou, N.; Banyko, J.; Brdicka, R.; Barbujani, G. et al. (2004). "Y chromosomal haplogroup J as a signature of the post-neolithic colonization of Europe". Human Genetics 115 (5): 357–71. doi:10.1007/s00439-004-1168-9. PMID 15322918.
- Eaaswarkhanth, Muthukrishnan; Haque, Ikramul; Ravesh, Zeinab; Romero, Irene Gallego; Meganathan, Poorlin Ramakodi; Dubey, Bhawna; Khan, Faizan Ahmed; Chaubey, Gyaneshwer et al. (March 2010). "Traces of sub-Saharan and Middle Eastern lineages in Indian Muslim populations". European Journal of Human Genetics 18 (3): 354–363. doi:10.1038/ejhg.2009.168. PMID 19809480.
- El-Sibai, M.; Platt, D. E.; Haber, M.; Xue, Y.; Youhanna, S. C.; Wells, R. S.; Izaabel, H.; Sanyoura, M. F. et al. (2009). "Geographical Structure of the Y-chromosomal Genetic Landscape of the Levant: A coastal-inland contrast". Annals of Human Genetics 73 (6): 568–81. doi:10.1111/j.1469-1809.2009.00538.x. PMID 19686289.
- Fadhlaoui-Zid, Karima (2014). "Sousse: extreme genetic heterogeneity in North Africa". Journal of Human Genetics 60 (1): 41–49. doi:10.1038/jhg.2014.99. PMID 25471516.
- Flores, Carlos; Maca-Meyer, Nicole; González, Ana M; Oefner, Peter J; Shen, Peidong; Pérez, Jose A; Rojas, Antonio; Larruga, Jose M et al. (October 2004). "Reduced genetic structure of the Iberian peninsula revealed by Y-chromosome analysis: implications for population demography". European Journal of Human Genetics 12 (10): 855–863. doi:10.1038/sj.ejhg.5201225. PMID 15280900.
- Gonçalves, Rita; Freitas, Ana; Branco, Marta; Rosa, Alexandra; Fernandes, Ana T.; Zhivotovsky, Lev A.; Underhill, Peter A.; Kivisild, Toomas et al. (2005). "Y-chromosome lineages from Portugal, Madeira and Açores record elements of Sephardim and Berber ancestry". Annals of Human Genetics 69 (Pt 4): 443–454. doi:10.1111/j.1529-8817.2005.00161.x. PMID 15996172.
- Hammer, Michael F.; Behar, Doron M.; Karafet, Tatiana M.; Mendez, Fernando L.; Hallmark, Brian; Erez, Tamar; Zhivotovsky, Lev A.; Rosset, Saharon et al. (2009). "Extended Y chromosome haplotypes resolve multiple and unique lineages of the Jewish priesthood". Human Genetics 126 (5): 707–17. doi:10.1007/s00439-009-0727-5. PMID 19669163.
- Jobling, Mark A.; Tyler-Smith, Chris (2000). "New uses for new haplotypes". Trends in Genetics 16 (8): 356–62. doi:10.1016/S0168-9525(00)02057-6. PMID 10904265.
- Karafet, T. M.; Mendez, F. L.; Meilerman, M. B.; Underhill, P. A.; Zegura, S. L.; Hammer, M. F. (2008). "New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree". Genome Research 18 (5): 830–8. doi:10.1101/gr.7172008. PMID 18385274.
- Luca, F.; Di Giacomo, F.; Benincasa, T.; Popa, L.O.; Banyko, J.; Kracmarova, A.; Malaspina, P.; Novelletto, A. et al. (2007). "Y-chromosomal variation in the Czech Republic". American Journal of Physical Anthropology 132 (1): 132–9. doi:10.1002/ajpa.20500. PMID 17078035.
- Luis, J; Rowold, D; Regueiro, M; Caeiro, B; Cinnioglu, C; Roseman, C; Underhill, P; Cavallisforza, L et al. (2004). "The Levant versus the Horn of Africa: Evidence for Bidirectional Corridors of Human Migrations". The American Journal of Human Genetics 74 (3): 532–44. doi:10.1086/382286. PMID 14973781.
- Martinez, Laisel; Underhill, Peter A; Zhivotovsky, Lev A; Gayden, Tenzin; Moschonas, Nicholas K; Chow, Cheryl-Emiliane T; Conti, Simon; Mamolini, Elisabetta et al. (April 2007). "Paleolithic Y-haplogroup heritage predominates in a Cretan highland plateau". European Journal of Human Genetics 15 (4): 485–493. doi:10.1038/sj.ejhg.5201769. PMID 17264870.
- "Human Y-chromosome short tandem repeats: A tale of acculturation and migrations as mechanisms for the diffusion of agriculture in the Balkan Peninsula". American Journal of Physical Anthropology 142 (3): 380–390. July 2010. doi:10.1002/ajpa.21235. PMID 20091845.
- Nasidze, I.; Ling, E. Y. S.; Quinque, D.; Dupanloup, I.; Cordaux, R.; Rychkov, S.; Naumova, O.; Zhukova, O. et al. (2004). "Mitochondrial DNA and Y-Chromosome Variation in the Caucasus". Annals of Human Genetics 68 (3): 205–21. doi:10.1046/j.1529-8817.2004.00092.x. PMID 15180701.
- Nebel, Almut; Filon, Dvora; Brinkmann, Bernd; Majumder, Partha P.; Faerman, Marina; Oppenheim, Ariella (November 2001). "The Y Chromosome Pool of Jews as Part of the Genetic Landscape of the Middle East". The American Journal of Human Genetics 69 (5): 1095–1112. doi:10.1086/324070. PMID 11573163.
- Onofri, Valerio; Alessandrini, Federica; Turchi, Chiara; Pesaresi, Mauro; Tagliabracci, Adriano (2008). "Y-chromosome markers distribution in Northern Africa: High-resolution SNP and STR analysis in Tunisia and Morocco populations". Forensic Science International: Genetics Supplement Series 1: 235–236. doi:10.1016/j.fsigss.2007.10.173.
- "High-resolution phylogenetic analysis of southeastern Europe traces major episodes of paternal gene flow among Slavic populations". Mol. Biol. Evol. 22 (10): 1964–75. October 2005. doi:10.1093/molbev/msi185. PMID 15944443.
- Robino, C.; Crobu, F.; Di Gaetano, C.; Bekada, A.; Benhamamouch, S.; Cerutti, N.; Piazza, A.; Inturri, S. et al. (2008). "Analysis of Y-chromosomal SNP haplogroups and STR haplotypes in an Algerian population sample". International Journal of Legal Medicine 122 (3): 251–255. doi:10.1007/s00414-007-0203-5. PMID 17909833.
- Semino, Ornella; Magri, Chiara; Benuzzi, Giorgia; Lin, Alice A.; Al-Zahery, Nadia; Battaglia, Vincenza; Maccioni, Liliana; Triantaphyllidis, Costas et al. (2004). "Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the neolithization of Europe and later migratory events in the Mediterranean area". American Journal of Human Genetics 74 (5): 1023–1034. doi:10.1086/386295. PMID 15069642.
- Shen, Peidong; Lavi, Tal; Kivisild, Toomas; Chou, Vivian; Sengun, Deniz; Gefel, Dov; Shpirer, Issac; Woolf, Eilon et al. (2004). "Reconstruction of patrilineages and matrilineages of Samaritans and other Israeli populations from Y-Chromosome and mitochondrial DNA sequence Variation". Human Mutation 24 (3): 248–60. doi:10.1002/humu.20077. PMID 15300852.
- Xue, Yali; Zerjal, Tatiana; Bao, Weidong; Zhu, Suling; Shu, Qunfang; Xu, Jiujin; Du, Ruofu; Fu, Songbin et al. (1 April 2006). "Male Demography in East Asia: A North–South Contrast in Human Population Expansion Times". Genetics 172 (4): 2431–2439. doi:10.1534/genetics.105.054270. PMID 16489223.
- Zalloua, Pierre A.; Platt, Daniel E.; El Sibai, Mirvat; Khalife, Jade; Makhoul, Nadine; Haber, Marc; Xue, Yali; Izaabel, Hassan et al. (2008). "Identifying Genetic Traces of Historical Expansions: Phoenician Footprints in the Mediterranean". American Journal of Human Genetics 83 (5): 633–642. doi:10.1016/j.ajhg.2008.10.012. PMID 18976729.
Thesis and Dissertations
- Varzari, Alexander (2006). Population History of the Dniester-Carpathians: Evidence from Alu Insertion and Y-Chromosome Polymorphisms (PDF) (Thesis). München, University. OCLC 180859661.
- Yunusbaev, Bayazit Bulatovich (2006). ПОПУЛЯЦИОННО-ГЕНЕТИЧЕСКОЕ ИССЛЕДОВАНИЕ НАРОДОВ ДАГЕСТАНА ПО ДАННЫМ О ПОЛИМОРФИЗМЕ У-ХРОМОСОМЫ И АТД-ИНСЕРЦИЙ [Population-genetic study of the peoples of Dagestan on the data on Y-chromosome and ATD-insertion polymorphism] (PDF) (PhD). Moscow: Russian Academy of Sciences. Archived from the original (PDF) on 5 February 2007.
Blogs
- Dienekes (2009). "Middle Eastern and Sub-Saharan lineages in Indian Muslim populations". http://dienekes.blogspot.com/2009/10/middle-eastern-and-sub-saharan-lineages.html.
Mailing Lists
- Aburto, Alfred A (2006). "Y haplogroup J in Iran" (Mailing list). Archived from the original on 13 October 2012. Retrieved 3 January 2013.
Websites
- Krahn; FTDNA (2003). "Genomic Research Center Draft Tree (AKA Y-TRee)". http://ytree.ftdna.com/index.php?name=Draft&parent=root.
Sources for conversion tables
- Capelli, Cristian; Wilson, James F.; Richards, Martin; Stumpf, Michael P.H. et al. (February 2001). "A Predominantly Indigenous Paternal Heritage for the Austronesian-Speaking Peoples of Insular Southeast Asia and Oceania". The American Journal of Human Genetics 68 (2): 432–443. doi:10.1086/318205. PMID 11170891.
- Hammer, Michael F.; Karafet, Tatiana M.; Redd, Alan J.; Jarjanazi, Hamdi et al. (1 July 2001). "Hierarchical Patterns of Global Human Y-Chromosome Diversity". Molecular Biology and Evolution 18 (7): 1189–1203. doi:10.1093/oxfordjournals.molbev.a003906. PMID 11420360.
- Jobling, Mark A.; Tyler-Smith, Chris (2000), "New uses for new haplotypes", Trends in Genetics 16 (8): 356–62, doi:10.1016/S0168-9525(00)02057-6, PMID 10904265
- Kaladjieva, Luba; Calafell, Francesc; Jobling, Mark A; Angelicheva, Dora et al. (February 2001). "Patterns of inter- and intra-group genetic diversity in the Vlax Roma as revealed by Y chromosome and mitochondrial DNA lineages". European Journal of Human Genetics 9 (2): 97–104. doi:10.1038/sj.ejhg.5200597. PMID 11313742.
- Karafet, Tatiana; Xu, Liping; Du, Ruofu; Wang, William et al. (September 2001). "Paternal Population History of East Asia: Sources, Patterns, and Microevolutionary Processes". The American Journal of Human Genetics 69 (3): 615–628. doi:10.1086/323299. PMID 11481588.
- Semino, O.; Passarino, G; Oefner, PJ; Lin, AA et al. (2000), "The Genetic Legacy of Paleolithic Homo sapiens sapiens in Extant Europeans: A Y Chromosome Perspective", Science 290 (5494): 1155–9, doi:10.1126/science.290.5494.1155, PMID 11073453, Bibcode: 2000Sci...290.1155S
- Su, Bing; Xiao, Junhua; Underhill, Peter; Deka, Ranjan et al. (December 1999). "Y-Chromosome Evidence for a Northward Migration of Modern Humans into Eastern Asia during the Last Ice Age". The American Journal of Human Genetics 65 (6): 1718–1724. doi:10.1086/302680. PMID 10577926.
- Underhill, Peter A.; Shen, Peidong; Lin, Alice A.; Jin, Li et al. (November 2000). "Y chromosome sequence variation and the history of human populations". Nature Genetics 26 (3): 358–361. doi:10.1038/81685. PMID 11062480.
Further reading
- yJdb: the Y-haplogroup J database haplotypes of haplogroup J.
- [1]
- Haplogroup J subclades at International Society of Genetic Genealogy
- Sanchez, Juan J; Hallenberg, Charlotte; Børsting, Claus; Hernandez, Alexis; Gorlin, RJ (2005). "High frequencies of Y chromosome lineages characterized by E3b1, DYS19-11, DYS392-12 in Somali males". European Journal of Human Genetics 13 (7): 856–66. doi:10.1038/sj.ejhg.5201390. PMID 15756297.
- "Polarity and temporality of high-resolution y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists". Am. J. Hum. Genet. 78 (2): 202–21. February 2006. doi:10.1086/499411. PMID 16400607.
Phylogenetic notes
- ↑ ISOGG Y-DNA Haplogroup J and its Subclades - 2016 (2 February 2016).
- ↑ This table shows the historic names for J-M304 (a.k.a. J-P209, and J-12f2.1) in published peer reviewed literature. Note that in Semino 2000 Eu09 is a subclade of Eu10 and in Karafet 2001 24 is a subclade of 23.
YCC 2002/2008 (Shorthand) J-M304
(a.k.a. J-12f2.1 or J-P209)Jobling and Tyler-Smith 2000 9 Underhill 2000 VI Hammer 2001 Med Karafet 2001 23 Semino 2000 Eu10 Su 1999 H4 Capelli 2001 B YCC 2002 (Longhand) J* YCC 2005 (Longhand) J YCC 2008 (Longhand) J YCC 2010r (Longhand) J - ↑ This table shows the historic names for J-M267 and its earlier discovered and named subclade J-M62 in published peer reviewed literature.
YCC 2002/2008 (Shorthand) J-M267 J-M62 Jobling and Tyler-Smith 2000 - 9 Underhill 2000 - VI Hammer 2001 - Med Karafet 2001 - 23 Semino 2000 - Eu10 Su 1999 - H4 Capelli 2001 - B YCC 2002 (Longhand) - J1 YCC 2005 (Longhand) J1 J1a YCC 2008 (Longhand) J1 J1a YCC 2010r (Longhand) J1 J1a - ↑ This table shows the historic names for J-M172 in published peer reviewed literature. Note that in Semino 2000 Eu09 is a subclade of Eu10 and in Karafet 2001 24 is a subclade of 23.
YCC 2002/2008 (Shorthand) J-M172 Jobling and Tyler-Smith 2000 9 Underhill 2000 VI Hammer 2001 Med Karafet 2001 24 Semino 2000 Eu9 Su 1999 H4 Capelli 2001 B YCC 2002 (Longhand) J2* YCC 2005 (Longhand) J2 YCC 2008 (Longhand) J2 YCC 2010r (Longhand) J2
External links
Phylogenetic tree and Distribution Maps of Y-DNA haplogroup J
Others
- Youtube Haplogroup J2 Channel
- Spread of Haplogroup J, from National Geographic
- British Isles DNA Project
 |

