Biology:L-Ribonucleic acid aptamer
An L-ribonucleic acid aptamer (L-RNA aptamer, trade name Spiegelmer) is an RNA-like molecule built from L-ribose units.[1] It is an artificial oligonucleotide named for being a mirror image of natural oligonucleotides. L-RNA aptamers are a form of aptamers. Due to their L-nucleotides, they are highly resistant to degradation by nucleases.[2] L-RNA aptamers are considered potential drugs and are currently being tested in clinical trials.
Features
Chemical properties
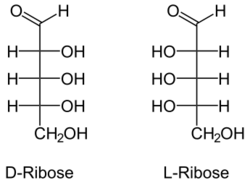
L-RNA aptamers, built using L-ribose, are the enantiomers of natural oligonucleotides, which are made with D-ribose. Nucleic acid aptamers, including L-RNA aptamers, contain adenosine monophosphate, guanosine monophosphate, cytidine monophosphate, uridine monophosphate, a phosphate group, a nucleobase and a ribose sugar.
Biological characteristics
Like other aptamers, L-RNA aptamers are able to bind molecules such as peptides, proteins, and substances of low molecular weight. The affinity of L-RNA aptamers to their target molecules often lies in the pico to nanomolar range and is thus comparable to antibodies.[clarification needed][3]
L-RNA aptamers themselves have low antigenicity. In contrast to other aptamers, L-RNA aptamers have high stability in blood serum, since they are less susceptible to be cleaved hydrolytically by enzymes.[4] They are excreted by the kidneys in a short time due to their low molar mass (which is below the renal threshold).
L-RNA aptamers modified with a higher molar mass, such as PEGylated L-RNA aptamers, show a prolonged plasma half-life.
Production
Unlike other aptamers, L-RNA aptamers are not directly made using systematic evolution of ligands by exponential enrichment (SELEX), as L-nucleic acids are not amenable to enzymatic methods, such as polymerase chain reaction (PCR), used in SELEX. Therefore, the selection is done with mirrored target molecules.
Reflection of the target molecule
The first step is the production of the target's enantiomer. In the case of peptides and small proteins that are produced synthetically, an enantiomer is made using synthetic D-amino acids. If the target is a larger protein molecule, beyond synthetic abilities, the enantiomer of an epitope is produced.[4]
SELEX
Conventional (up to 1016 different oligonucleotides) existing molecule library serves as a starting point for the subsequent SELEX process.[clarification needed] Selection, separation, and amplification using the mirror image of the target molecule is performed.
Sequencing and synthesis
The sequence of the oligonucleotide selected using SELEX is determined with the help of DNA sequencing. This information is used for the synthesis of the oligonucleotide's enantiomer, the L-RNA aptamer, using L-nucleotides.
Use
L-RNA aptamers have been obtained for the chemokines CCL2 and CXCL12, the complement components C5a and ghrelin. They are currently in preclinical or clinical development. Proof-of-concept for an anti-CCL2/MCP-1 L-RNA aptamers has recently been demonstrated in diabetic nephropathy patients.[2] They can also be used as diagnostic agents.[4]
References
- ↑ Helmling, S.H.; Eulberg, D.E.; Maasch, C.M.; Buchner, K.B.; Klussmann, S.K. (July 2003). "RNA-Spiegelmers: a new substance class to efficiently inhibit peptide hormones". European Journal of Biochemistry 271 (S1). doi:10.1111/j.1474-3833.2004.4119.x.
- ↑ 2.0 2.1 "Turning mirror-image oligonucleotides into drugs: the evolution of Spiegelmer therapeutics". Drug Discovery Today 20 (1): 147–155. January 2015. doi:10.1016/j.drudis.2014.09.004. PMID 25236655.
- ↑ Wlotzka, Britta; Leva, Susanne; Eschgfäller, Bernd; Burmeister, Jens; Kleinjung, Frank; Kaduk, Christine; Muhn, Peter; Hess-Stumpp, Holger et al. (June 2002). "In vivo properties of an anti-GnRH Spiegelmer: an example of an oligonucleotide-based therapeutic substance class". Proceedings of the National Academy of Sciences of the United States of America 99 (13): 8898–902. doi:10.1073/pnas.132067399. PMID 12070349. Bibcode: 2002PNAS...99.8898W.
- ↑ 4.0 4.1 4.2 "Mirror-image RNA that binds D-adenosine". Nat. Biotechnol. 14 (9): 1112–5. September 1996. doi:10.1038/nbt0996-1112. PMID 9631061.
 |