Biology:Metallo-beta-lactamase protein fold
| Metallo-beta-lactamase superfamily | |||||||||
|---|---|---|---|---|---|---|---|---|---|
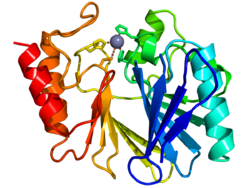 Cartoon diagram of the crystallographic structure of a zinc metallo-beta-lactamase from bacillus cereus (N-terminus = blue, C-terminus = red). The catalytic zinc ion is depicted as a grey sphere. | |||||||||
| Identifiers | |||||||||
| Symbol | Lactamase_B | ||||||||
| Pfam | PF00753 | ||||||||
| InterPro | IPR001279 | ||||||||
| SCOP2 | 1bmc / SCOPe / SUPFAM | ||||||||
| |||||||||
The metallo-β-lactamase (MBL) superfamily[1] constitutes a group of proteins found in all domains of life that share a characteristic αββα fold with the ability to bind transition metal ions. Such metal binding sites may have divalent transition metal ions like Zn(II), Fe(II)/Fe(III) and Mn(II), and are located at the bottom of a wide cleft able to accommodate diverse substrates. The name was adopted after the first members of the superfamily to be studied experimentally: a group of zinc-dependent hydrolytic enzymes conferring bacterial resistance to β-lactam antibiotics. These zinc-β-lactamases (ZBLs) inactivate β-lactam antibiotics through hydrolysis of the β-lactam ring. Early studies on MBLs were conducted on the enzyme βLII isolated from strain 569/H/9 of Bacillus cereus. It was named βLII because it was the second β-lactamase shown to be produced by the bacterium; the first one, βLI, was a non-metallic β-lactamase, i.e., insensitive to inhibition with EDTA (βLII was renamed BcII over time).
Low-resolution X-ray crystallographic analyses published in 1995,[2] disclosed the new αββα fold that would become the hallmark of the MBL superfamily, along with a single Zn(II) ion bound to a three-histidine motif, resembling the active site typical of carbonic anhydrases. Thus, BcII and ZBLs in general were thought to use a single Zn(II) ion to activate a water molecule for hydrolysis, analogous to the mechanism by which carbonic anhydrases hydrate carbon dioxide into bicarbonate. This belief was soon debunked when the structure of Bacteroides fragilis ZBL, CcrA, was published, showing an additional Zn(II) ion next to the previous one. The second zinc was coordinated to nearby Asp, Cys and His residues.[3] Besides, the second metal ion was later found in Bacillus cereus ZBL too,[4] starting a decade-long controversy regarding the role of each zinc ion. Later on, it was found that monometallic ZBLs are rather exceptional and the antibiotic inactivation reaction requires two Zn(II) ions.[5] Similarly, it has been demonstrated that zinc chelators can inhibit the hydrolytic activity of metallo-β-lactamases against β-lactam antibiotics, restoring the activity of the latter.[6]
Metallo-beta-lactamases are important enzymes because they are involved in the breakdown of antibiotics by antibiotic-resistant bacteria.[7] It is unclear whether metallo-beta-lactamase activity evolved once or twice within the superfamily; if twice, this would suggest structural exaptation.[8] Proteins belonging to the MBL superfamily usually combine at least one MBL domain with additional domains that provide different functions, such as substrate recognition or binding to other polypeptides, in a modular fashion. Thus, MBL superfamily members grasp the metal-assisted water-activation ability of the MBL domain in order to perform a wide variety of hydrolytic reactions. Such diversity is often expanded by mutations around the metal-binding site in order to bind different metal ions. Indeed, those MBLs that bind Fe(II)/Fe(III) are often redox active due to the ability to perform one-electron redox reactions.
Early attempts to systematically classify all members of the MBL superfamily were conducted in 1999 by Aravind,[9] who showed that many other proteins display the αββα typical of MBLs. These observations were updated in 2001 by Daiyasu et al. who defined at least 16 families within the MBL superfamily.[10] These proteins include thiolesterases, members of the glyoxalase II family, that catalyse the hydrolysis of S-D-lactoyl-glutathione to form glutathione and D-lactic acid and a competence protein that is essential for natural transformation in Neisseria gonorrhoeae and could be a transporter involved in DNA uptake. Except for the competence protein these proteins bind two zinc ions per molecule as cofactor. Currently, at least one hundred proteins have been shown to contain an αββα domain using X-ray crystallography, whereas the whole MBL superfamily includes about half a million members.
See also
- New Delhi metallo-beta-lactamase
References
- ↑ "Visualizing the superfamily of metallo-β-lactamases through sequence similarity network neighborhood connectivity analysis". Heliyon 7 (1): e05867. 2021. doi:10.1016/j.heliyon.2020.e05867. PMID 33426353.
- ↑ "The 3-D structure of a zinc metallo-beta-lactamase from Bacillus cereus reveals a new type of protein fold". EMBO Journal 14 (20): 4914–21. 1995. doi:10.1002/j.1460-2075.1995.tb00174.x. PMID 7588620.
- ↑ "Crystal structure of the wide-spectrum binuclear zinc beta-lactamase from Bacteroides fragilis". Structure 4 (7): 823–36. 1996. doi:10.1016/s0969-2126(96)00089-5. PMID 8805566.
- ↑ "Spectroscopic characterization of a binuclear metal site in Bacillus cereus beta-lactamase II". Biochemistry 37 (28): 10137–80. 1998. doi:10.1021/bi980309j. PMID 9665723.
- ↑ "Metallo-β-lactamases withstand low Zn(II) conditions by tuning metal-ligand interactions". Nature Chemical Biology 8 (8): 698–700. 2012. doi:10.1038/nchembio.1005. PMID 22729148.
- ↑ Principe, Luigi; Vecchio, Graziella; Sheehan, Gerard; Kavanagh, Kevin; Morroni, Gianluca; Viaggi, Valentina; di Masi, Alessandra; Giacobbe, Daniele Roberto et al. (2020-10-01). "Zinc Chelators as Carbapenem Adjuvants for Metallo-β-Lactamase-Producing Bacteria: In Vitro and In Vivo Evaluation" (in en). Microbial Drug Resistance 26 (10): 1133–1143. doi:10.1089/mdr.2020.0037. ISSN 1076-6294. PMID 32364820. https://www.liebertpub.com/doi/10.1089/mdr.2020.0037.
- ↑ Shaw, Robert W; Kim, Sung-kun (November 2008). "Inhibition of metallo-β-lactamase". http://www.freepatentsonline.com/7456274.html.
- ↑ "One origin for metallo-beta-lactamase activity, or two? An investigation assessing a diverse set of reconstructed ancestral sequences based on a sample of phylogenetic trees". J. Mol. Evol. 79 (3–4): 117–29. 2014. doi:10.1007/s00239-014-9639-7. PMID 25185655. Bibcode: 2014JMolE..79..117A.
- ↑ "An evolutionary classification of the metallo-beta-lactamase fold proteins". In Silico Biology 1 (2): 69–91. 1999. PMID 11471246.
- ↑ "Expansion of the zinc metallo-hydrolase family of the beta-lactamase fold". FEBS Letters 503 (1): 1–6. 2001. doi:10.1016/s0014-5793(01)02686-2. PMID 11513844.
 |

