Biology:N-acylglucosamine 2-epimerase
| N-acylglucosamine 2-epimerase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
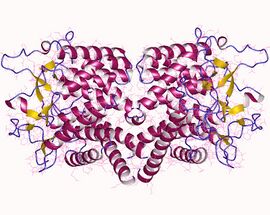 N-acylglucosamine 2-epimerase dimer, Sus scrofa | |||||||||
| Identifiers | |||||||||
| EC number | 5.1.3.8 | ||||||||
| CAS number | 37318-34-6 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
In enzymology, a N-acylglucosamine 2-epimerase (EC 5.1.3.8) is an enzyme that catalyzes the chemical reaction
- N-acyl-D-glucosamine N-acyl-D-mannosamine
Hence, this enzyme has one substrate, N-acyl-D-glucosamine, and one product, N-acyl-D-mannosamine.
This enzyme belongs to the family of isomerases, specifically those racemases and epimerases acting on carbohydrates and derivatives. The systematic name of this enzyme class is N-acyl-D-glucosamine 2-epimerase. Other names in common use include acylglucosamine 2-epimerase, and N-acetylglucosamine 2-epimerase. This enzyme participates in aminosugars metabolism. It employs one cofactor, ATP.
Structural studies
As of late 2019, three structures have been solved for this class of enzymes, with the PDB accession codes 1FP3, 2GZ6, and 6F04. They show that the N-acylglucosamine 2-epimerase monomer folds as a barrel composed of α-helices, in a manner known as (α/α)6-barrel. The structures are presented as dimers, with the structures from Sus scrofa and Anabaena sp. CH1 having a different organization than the structure from Nostoc sp. KJV10.
References
- "The sialic acids. V. N-Acyl-D-glucosamine 2-epimerase". The Journal of Biological Chemistry 240: 1531–6. April 1965. doi:10.1016/S0021-9258(18)97468-5. PMID 14285488.
 |

