Biology:OBPgp279
| OBPgp279 | |||||||
|---|---|---|---|---|---|---|---|
 Predicted structure of OPBgp279 from Phyre2. Homology model based on 4OK7. | |||||||
| Identifiers | |||||||
| Organism | |||||||
| Symbol | OBP_279 | ||||||
| UniProt | G9I9L2 | ||||||
| |||||||
OBPgp279 (OBP genome protein 279) is an endolysin that hydrolyzes peptidoglycan, a major constituent in bacterial membrane.[1] OBPgp279 is found in Pseudomonas fluorescens phage OBP, which belongs in the Myoviridae family of bacteriophages. Because of its role in hydrolyzing the peptidoglycan layer, OBPgp279 is a key enzyme in the lytic cycle of the OBP bacteriophage; it allows the bacteriophage to lyse its host internally to escape. Unlike other endolysins, OBPgp279 does not rely on holins to perforate the inner bacterial membrane in order to reach the peptidoglycan layer.[1][2] Although OBPgp279 is not a well-studied enzyme, it has garnered interest as a potential antibacterial protein due to its activity against multidrug-resistant gram-negative bacteria.[3]
Predicted enzyme mechanism
The mechanism of OBPgp279 is predicted to be part of glycoside hydrolase family 19 (GH19) due to the presence of a conserved sequence motif (general sequence motif = [FHY]-G-R-G-[AP]-ζ-Q-[IL]-[ST]-[FHYW]-[HN]-[FY]-[NY], ζ= hydrophilic amino acid) in the catalytic domain of the enzyme.[1][5] GH19 is a group of endo-acting enzymes that hydrolyzes the glycosidic bonds of β-1,4-linked N-Acetylglucosamine (GlcNAc) typically within chitin; however, some enzymes in this family also demonstrate lysozyme activity.[citation needed]
Since bacterial cell walls do not contain chitin, OBPgp279 hydrolyzes β-1,4-linked GlcNAc in peptidoglycan.[2] The hydrolysis of β-1,4-linked GlcNAc is catalyzed by two glutamate residues in the active side, one acting as a general acid, and another acting as a general base.[6]
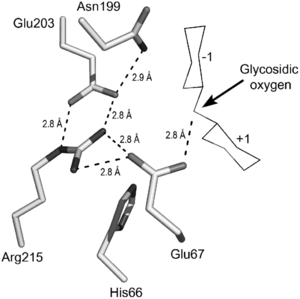
There is limited detail on the catalytic mechanism of OBPgp279. However, because OBPgp279 is part of GH19, it is possible to infer the structure of OBPgp279 binding to peptidoglycan from the substrate binding of glycoside hydrolase family 19.[4] The figure on the right shows an example of a glycoside hydrolase family 19 binding to chitin. OBPgp279 most likely has a similar active site, but it binds to peptidoglycan instead of chitin.
Due to the activity of OBPgp279 on β-1,4-linked GlcNAc, it is likely that OBPgp279 is a N-acetylmuramidase (lysozyme-like) endolysin which hydrolyzes the sugar backbone component of the peptidoglycan on the reducing side of GlcNAc.[7]
Enzyme domain organization
The catalytic domain of the enzyme resides on the C-terminal region of the enzyme. OBPgp279 is also predicted to contain peptidoglycan binding domains; since OBPgp279 contains two peptidoglycan binding domain motifs in its N-terminal region (general sequence motif = D-G-(Pho)2-G-K/N-G/N-T, Pho = hydrophobic amino acid), it likely contains two peptidoglycan binding domains as shown in the schematic drawing below.[1][8]

Application
OBPgp279 has gained attention as a potential antibiotic because of the increasing prevalence of multidrug resistant gram-negative bacteria.[9] Typically, most endolysins rely on holin to reach the peptidoglycan layer of gram-negative bacteria. This limits their efficacy as standalone antibiotics; without holins or any membrane permeabilizers, they have low membrane penetration. In contrast, OBPgp279 is capable of penetrating the bacterial outer membrane to reach the peptidoglycan layer without needing holins or any membrane permeabilizers.[2]
In addition, unlike most endolysins, OBPgp279 demonstrates high efficacy against a variety of bacteria instead of being species-specific.[1] Compared to small molecule antibiotics, OBPgp279 also has the advantages of a low chance of bacterial resistance, specificity towards pathogen-causing bacteria, and efficacy on mucosal surfaces.[10]
In 2013, KU Leuven professor Rob Lavigne modified OBPgp279 by adding a polycationic nonapeptide to its C-terminus, thereby improving its efficacy as an antibiotic.[9] The addition of the polycationic nonapeptide improved OBPgp279’s log reduction against various gram-negative and gram-positive bacteria by as much as three logs. This improvement is likely because of increased outer membrane penetration, in turn caused by favorable electrostatic interaction between the cationic nonapeptide and the negatively charged bacterial membrane.[11] The engineered OBPgp279, along with the engineering technology platform, is currently owned by Boehringer Ingelheim Vetmedica.[12]
The major drawback of working with OBPgp279 and other endolysins is their immunogenicity. Although studies have shown that antibodies do not affect the efficacy of endolysins in animal models, immunogenicity will need to be monitored if OBPgp279 is pursued for medical use.[13]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 "Characterization of modular bacteriophage endolysins from Myoviridae phages OBP, 201φ2-1 and PVP-SE1". PLOS ONE 7 (5): e36991. 2012-05-15. doi:10.1371/journal.pone.0036991. PMID 22615864. Bibcode: 2012PLoSO...736991W.
- ↑ 2.0 2.1 2.2 "Endolysins as antimicrobials". Advances in Virus Research. Bacteriophages, Part B (Academic Press) 83: 299–365. 2012-01-01. doi:10.1016/b978-0-12-394438-2.00007-4. ISBN 9780123944382. PMID 22748813.
- ↑ "The Rockefeller University » Hospital Centennial". http://centennial.rucares.org/index.php?page=Phage_Lysins.
- ↑ 4.0 4.1 "Structure and function of enzymes acting on chitin and chitosan". Biotechnology & Genetic Engineering Reviews 27 (1): 331–66. 2010-01-01. doi:10.1080/02648725.2010.10648156. PMID 21415904.
- ↑ "Evolution, homology conservation, and identification of unique sequence signatures in GH19 family chitinases". Journal of Molecular Evolution 70 (5): 466–78. May 2010. doi:10.1007/s00239-010-9345-z. PMID 20480157. Bibcode: 2010JMolE..70..466U.
- ↑ "X-ray structure of papaya chitinase reveals the substrate binding mode of glycosyl hydrolase family 19 chitinases". Biochemistry 47 (32): 8283–91. August 2008. doi:10.1021/bi800655u. PMID 18636748.
- ↑ "Bacteriophage endolysins as novel antimicrobials". Future Microbiology 7 (10): 1147–71. October 2012. doi:10.2217/fmb.12.97. PMID 23030422.
- ↑ "Muralytic activity and modular structure of the endolysins of Pseudomonas aeruginosa bacteriophages phiKZ and EL". Molecular Microbiology 65 (5): 1334–44. September 2007. doi:10.1111/j.1365-2958.2007.05870.x. PMID 17697255.
- ↑ 9.0 9.1 "Engineered endolysin-based "Artilysins" to combat multidrug-resistant gram-negative pathogens". mBio 5 (4): e01379-14. July 2014. doi:10.1128/mBio.01379-14. PMID 24987094.
- ↑ "Bacteriophage endolysins: a novel anti-infective to control Gram-positive pathogens". International Journal of Medical Microbiology 300 (6): 357–62. August 2010. doi:10.1016/j.ijmm.2010.04.002. PMID 20452280.
- ↑ "The bacterial cell envelope". Cold Spring Harbor Perspectives in Biology 2 (5): a000414. May 2010. doi:10.1101/cshperspect.a000414. PMID 20452953.
- ↑ "Boehringer Ingelheim Vetmedica Acquires Licensing Rights for Artilysin® Technology from Lysando AG | Business Wire". 2014-08-12. http://www.businesswire.com/news/home/20140812005343/en/Boehringer-Ingelheim-Vetmedica-Acquires-Licensing-Rights-Artilysin%2525C2%2525AE.
- ↑ "Bacteriophage lysins as effective antibacterials". Current Opinion in Microbiology 11 (5): 393–400. October 2008. doi:10.1016/j.mib.2008.09.012. PMID 18824123.
 |