Biology:Glycoside hydrolase family 19
| Glycosyl hydrolases family 19 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 the refined crystal structure of an endochitinase from hordeum vulgare l. seeds to 1.8 angstroms resolution | |||||||||
| Identifiers | |||||||||
| Symbol | Glyco_hydro_19 | ||||||||
| Pfam | PF00182 | ||||||||
| Pfam clan | CL0037 | ||||||||
| InterPro | IPR000726 | ||||||||
| PROSITE | PDOC00620 | ||||||||
| SCOP2 | 2baa / SCOPe / SUPFAM | ||||||||
| CAZy | GH19 | ||||||||
| CDD | cd00325 | ||||||||
| |||||||||
In molecular biology, Glycoside hydrolase family 19 is a family of glycoside hydrolases EC 3.2.1., which are a widespread group of enzymes that hydrolyse the glycosidic bond between two or more carbohydrates, or between a carbohydrate and a non-carbohydrate moiety. A classification system for glycoside hydrolases, based on sequence similarity, has led to the definition of >100 different families.[1][2][3] This classification is available on the CAZy web site,[4][5] and also discussed at CAZypedia, an online encyclopedia of carbohydrate active enzymes.[6][7] y[ _]9
Glycoside hydrolase family 19 CAZY GH_19 comprises enzymes with only one known activity; chitinase (EC 3.2.1.14).
Chitinases[8] are enzymes that catalyze the hydrolysis of the beta-1,4-N-acetyl-D-glucosamine linkages in chitin polymers. Chitinases belong to glycoside hydrolase families 18 or 19.[9] Chitinases of family 19 (also known as classes IA or I and IB or II) are enzymes from plants that function in the defence against fungal and insect pathogens by destroying their chitin-containing cell wall. Class IA/I and IB/II enzymes differ in the presence (IA/I) or absence (IB/II) of a N-terminal chitin-binding domain. The catalytic domain of these enzymes consist of about 220 to 230 amino acid residues.
Active site
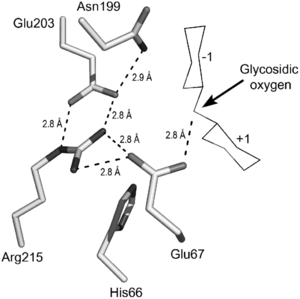
GH19 enzymes has a conserved sequence motif ([FHY]-G-R-G-[AP]-ζ-Q-[IL]-[ST]-[FHYW]-[HN]-[FY]-[NY], ζ= hydrophilic amino acid) in its active site.[11]
References
- ↑ "Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases". Proceedings of the National Academy of Sciences of the United States of America 92 (15): 7090–4. July 1995. doi:10.1073/pnas.92.15.7090. PMID 7624375. Bibcode: 1995PNAS...92.7090H.
- ↑ "Structures and mechanisms of glycosyl hydrolases". Structure 3 (9): 853–9. September 1995. doi:10.1016/S0969-2126(01)00220-9. PMID 8535779.
- ↑ "Updating the sequence-based classification of glycosyl hydrolases". The Biochemical Journal 316 ( Pt 2) (Pt 2): 695–6. June 1996. doi:10.1042/bj3160695. PMID 8687420.
- ↑ "Home" (in en). http://www.cazy.org/.
- ↑ "The carbohydrate-active enzymes database (CAZy) in 2013". Nucleic Acids Research 42 (Database issue): D490-5. January 2014. doi:10.1093/nar/gkt1178. PMID 24270786.
- ↑ "Glycoside Hydrolase Family 19" (in en). http://www.cazypedia.org/index.php/Glycoside_Hydrolase_Family_19.
- ↑ CAZypedia Consortium (December 2018). "Ten years of CAZypedia: a living encyclopedia of carbohydrate-active enzymes". Glycobiology 28 (1): 3–8. doi:10.1093/glycob/cwx089. PMID 29040563. https://hal.archives-ouvertes.fr/hal-01886461/file/Hehemann_2018_01.pdf.
- ↑ "What's new in chitinase research?". Experientia 48 (8): 701–16. August 1992. doi:10.1007/BF02124285. PMID 1516675.
- ↑ "A classification of glycosyl hydrolases based on amino acid sequence similarities". The Biochemical Journal 280 ( Pt 2) (2): 309–16. December 1991. doi:10.1042/bj2800309. PMID 1747104.
- ↑ "Structure and function of enzymes acting on chitin and chitosan". Biotechnology & Genetic Engineering Reviews 27 (1): 331–66. 2010-01-01. doi:10.1080/02648725.2010.10648156. PMID 21415904.
- ↑ "Evolution, homology conservation, and identification of unique sequence signatures in GH19 family chitinases". Journal of Molecular Evolution 70 (5): 466–78. May 2010. doi:10.1007/s00239-010-9345-z. PMID 20480157. Bibcode: 2010JMolE..70..466U.
 |

