Biology:Plant evolution

Plant evolution is the subset of evolutionary phenomena that concern plants. Evolutionary phenomena are characteristics of populations that are described by averages, medians, distributions, and other statistical methods. This distinguishes plant evolution from plant development, a branch of developmental biology which concerns the changes that individuals go through in their lives. The study of plant evolution attempts to explain how the present diversity of plants arose over geologic time. It includes the study of genetic change and the consequent variation that often results in speciation, one of the most important types of radiation into taxonomic groups called clades. A description of radiation is called a phylogeny and is often represented by type of diagram called a phylogenetic tree.
Evolutionary trends
Differences between plant and animal physiology and reproduction cause minor differences in how they evolve.
One major difference is the totipotent nature of plant cells, allowing them to reproduce asexually much more easily than most animals. They are also capable of polyploidy – where more than two chromosome sets are inherited from the parents. This allows relatively fast bursts of evolution to occur, for example by the effect of gene duplication. The long periods of dormancy that seed plants can employ also makes them less vulnerable to extinction, as they can "sit out" the tough periods and wait until more clement times to leap back to life.[1]
The effect of these differences is most profoundly seen during extinction events. These events, which wiped out between 6 and 62% of terrestrial animal families, had "negligible" effect on plant families.[2] However, the ecosystem structure is significantly rearranged, with the abundances and distributions of different groups of plants changing profoundly.[2] These effects are perhaps due to the higher diversity within families, as extinction – which was common at the species level – was very selective. For example, wind-pollinated species survived better than insect-pollinated taxa, and specialised species generally lost out.[2] In general, the surviving taxa were rare before the extinction, suggesting that they were generalists who were poor competitors when times were easy, but prospered when specialised groups became extinct and left ecological niches vacant.[2]
During embryogenesis, plants and animals pass through a phylotypic stage that evolved independently[3] and that causes a developmental constraint limiting morphological diversification.[4][5][6][7]
Polyploidy
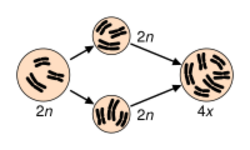
Polyploidy is pervasive in plants and some estimates suggest that 30–80% of living plant species are polyploid, and many lineages show evidence of ancient polyploidy (paleopolyploidy) in their genomes.[8][9][10] Huge explosions in angiosperm species diversity appear to have coincided with ancient genome duplications shared by many species.[11] 15% of angiosperm and 31% of fern speciation events are accompanied by ploidy increase.[12] Most polyploids display heterosis relative to their parental species, and may display novel variation or morphologies that may contribute to the processes of speciation and eco-niche exploitation.[9][13] The mechanisms leading to novel variation in newly formed allopolyploids may include gene dosage effects (resulting from more numerous copies of genome content), the reunion of divergent gene regulatory hierarchies, chromosomal rearrangements, and epigenetic remodeling, all of which affect gene content and/or expression levels.[14][15][16] Many of these rapid changes may contribute to reproductive isolation and speciation.
All eukaryotes probably have experienced a polyploidy event at some point in their evolutionary history. See paleopolyploidy. In many cases, these events can be inferred only through comparing sequenced genomes. Angiosperms have paleopolyploidy in their ancestry. Unexpected ancient genome duplications have recently been confirmed in mustard weed/thale cress (Arabidopsis thaliana) and rice (Oryza sativa).
Photosynthesis
Cyanobacteria and the evolution of photosynthesis
Cyanobacteria remained principal primary producers throughout the Proterozoic Eon (2500–543 Ma), in part because the redox structure of the oceans favored photoautotrophs capable of nitrogen fixation. [citation needed] Green algae joined blue-greens as major primary producers on continental shelves near the end of the Proterozoic, but only with the Mesozoic (251–65 Ma) radiations of dinoflagellates, coccolithophorids, and diatoms did primary production in marine shelf waters take modern form. Cyanobacteria remain critical to marine ecosystems as primary producers in oceanic gyres, as agents of biological nitrogen fixation, and, in modified form, as the plastids of marine algae.[17]
Symbiosis and the origin of chloroplasts
Chloroplasts have many similarities with cyanobacteria, including a circular chromosome, prokaryotic-type ribosomes, and similar proteins in the photosynthetic reaction center.[18][19] The endosymbiotic theory suggests that photosynthetic bacteria were acquired (by endocytosis) by early eukaryotic cells to form the first plant cells. Therefore, chloroplasts may be photosynthetic bacteria that adapted to life inside plant cells. Like mitochondria, chloroplasts still possess their own DNA, separate from the nuclear DNA of their plant host cells and the genes in this chloroplast DNA resemble those in cyanobacteria.[20] DNA in chloroplasts codes for redox proteins such as photosynthetic reaction centers. The CoRR hypothesis proposes that this Co-location is required for Redox Regulation.
Evolution of plant transcriptional regulation
Transcription factors and transcriptional regulatory networks play key roles in plant development and stress responses, as well as their evolution. During plant landing, many novel transcription factor families emerged and are preferentially wired into the networks of multicellular development, reproduction, and organ development, contributing to more complex morphogenesis of land plants.[21]
See also
References
- ↑ Willis, Charles G.; Baskin, Carol C.; Baskin, Jerry M.; Auld, Josh R.; Venable, D. Lawrence; Cavender‐Bares, Jeannine; Donohue, Kathleen; Rubio de Casas, Rafael et al. (July 2014). "The evolution of seed dormancy: environmental cues, evolutionary hubs, and diversification of the seed plants" (in en). New Phytologist 203 (1): 300–309. doi:10.1111/nph.12782. ISSN 0028-646X. PMID 24684268.
- ↑ 2.0 2.1 2.2 2.3 McElwain, J.C.; Punyasena, S.W. (2007). "Mass extinction events and the plant fossil record". Trends in Ecology & Evolution 22 (10): 548–557. doi:10.1016/j.tree.2007.09.003. PMID 17919771.
- ↑ Drost, Hajk-Georg; Janitza, Philipp; Grosse, Ivo; Quint, Marcel (2017). "Cross-kingdom comparison of the developmental hourglass". Current Opinion in Genetics & Development 45: 69–75. doi:10.1016/j.gde.2017.03.003. PMID 28347942.
- ↑ Irie, Naoki; Kuratani, Shigeru (2011-03-22). "Comparative transcriptome analysis reveals vertebrate phylotypic period during organogenesis" (in en). Nature Communications 2: 248. doi:10.1038/ncomms1248. ISSN 2041-1723. PMID 21427719. Bibcode: 2011NatCo...2..248I.
- ↑ Domazet-Lošo, Tomislav; Tautz, Diethard (2010-12-09). "A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns" (in en). Nature 468 (7325): 815–818. doi:10.1038/nature09632. ISSN 0028-0836. PMID 21150997. Bibcode: 2010Natur.468..815D.
- ↑ Quint, Marcel; Drost, Hajk-Georg; Gabel, Alexander; Ullrich, Kristian Karsten; Bönn, Markus; Grosse, Ivo (2012-10-04). "A transcriptomic hourglass in plant embryogenesis" (in en). Nature 490 (7418): 98–101. doi:10.1038/nature11394. ISSN 0028-0836. PMID 22951968. Bibcode: 2012Natur.490...98Q.
- ↑ Drost, Hajk-Georg; Gabel, Alexander; Grosse, Ivo; Quint, Marcel (2015-05-01). "Evidence for Active Maintenance of Phylotranscriptomic Hourglass Patterns in Animal and Plant Embryogenesis" (in en). Molecular Biology and Evolution 32 (5): 1221–1231. doi:10.1093/molbev/msv012. ISSN 0737-4038. PMID 25631928.
- ↑ "On the abundance of polyploids in flowering plants". Evolution 60 (6): 1198–206. June 2006. doi:10.1111/j.0014-3820.2006.tb01198.x. PMID 16892970.
- ↑ 9.0 9.1 "Plant speciation". Science 317 (5840): 910–4. August 2007. doi:10.1126/science.1137729. PMID 17702935. Bibcode: 2007Sci...317..910R.
- ↑ Otto SP (November 2007). "The evolutionary consequences of polyploidy". Cell 131 (3): 452–62. doi:10.1016/j.cell.2007.10.022. PMID 17981114.
- ↑ de Bodt et al. 2005
- ↑ "The frequency of polyploid speciation in vascular plants". Proc. Natl. Acad. Sci. U.S.A. 106 (33): 13875–9. August 2009. doi:10.1073/pnas.0811575106. PMID 19667210. Bibcode: 2009PNAS..10613875W.
- ↑ Comai L (November 2005). "The advantages and disadvantages of being polyploid". Nat. Rev. Genet. 6 (11): 836–46. doi:10.1038/nrg1711. PMID 16304599.
- ↑ "Understanding mechanisms of novel gene expression in polyploids". Trends Genet. 19 (3): 141–7. March 2003. doi:10.1016/S0168-9525(03)00015-5. PMID 12615008.
- ↑ "Mechanisms of genomic rearrangements and gene expression changes in plant polyploids". BioEssays 28 (3): 240–52. March 2006. doi:10.1002/bies.20374. PMID 16479580.
- ↑ Chen ZJ (2007). "Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids". Annu Rev Plant Biol 58: 377–406. doi:10.1146/annurev.arplant.58.032806.103835. PMID 17280525.
- ↑ Herrero A and Flores E (editor). (2008). The Cyanobacteria: Molecular Biology, Genomics and Evolution (1st ed.). Caister Academic Press. ISBN 978-1-904455-15-8.
- ↑ Douglas SE (1998). "Plastid evolution: origins, diversity, trends". Curr. Opin. Genet. Dev. 8 (6): 655–61. doi:10.1016/S0959-437X(98)80033-6. PMID 9914199.
- ↑ "The origin and establishment of the plastid in algae and plants". Annu. Rev. Genet. 41: 147–68. 2007. doi:10.1146/annurev.genet.41.110306.130134. PMID 17600460.
- ↑ "Genomics and chloroplast evolution: what did cyanobacteria do for plants?". Genome Biol. 4 (3): 209. 2003. doi:10.1186/gb-2003-4-3-209. PMID 12620099.
- ↑ Jin JP (July 2015). "An Arabidopsis transcriptional regulatory map reveals distinct functional and evolutionary features of novel transcription factors". Molecular Biology and Evolution 32 (7): 1767–1773. doi:10.1093/molbev/msv058. PMID 25750178.
External links
 |
