Biology:Postzygotic mutation
A postzygotic mutation (or post-zygotic mutation) is a change in an organism's genome that is acquired during its lifespan, instead of being inherited from its parent(s) through fusion of two haploid gametes. Mutations that occur after the zygote has formed can be caused by a variety of sources that fall under two classes: spontaneous mutations and induced mutations. How detrimental a mutation is to an organism is dependent on what the mutation is, where it occurred in the genome and when it occurred.[1]
Causes
Postzygotic changes to a genome can be caused by small mutations that affect a single base pair, or large mutations that affect entire chromosomes and are divided into two classes, spontaneous mutations and induced mutations.[2]
Spontaneous mutations
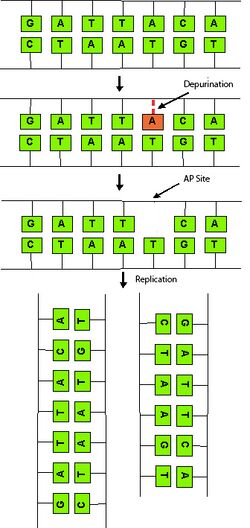
Most spontaneous mutations are the result of naturally occurring lesions to DNA and errors during DNA replication without direct exposure to an agent.[2] A few common spontaneous mutations are:
- Depurination – The loss of a purine (A or G) base to form an apurinic site. An apurinic site, also known as an AP site, is the location in a genetic sequence that does not contain a purine base. During replication, the affected double stranded DNA will produce one doubled-stranded daughter containing the missing purine, resulting in an unchanged sequence. The other strand will produce a shorter strand, missing the purine and its complementary base.[2][3]
- Deamination – The amine group on a base is changed to a keto group. This results in cytosine being changed to uracil and adenine being changed to hypoxanthine which can result in incorrect DNA replication and repair.[2][3]
- Tautomerization – The hydrogen atom on a nucleotide base is repositioned causing altered hydrogen bonding pattern and incorrect base pairing during replication.[2] For example, the keto tautomer of thymine normally pairs with adenine, however the enol tautomer of thymine can bind with guanine. This results in an incorrect base pair match. Similarly there are amino and imino tautomers of cytosine and adenine that can cause incorrect base pairing with other nucleotides.[4]
Induced mutations
Induced mutations are any lesions in DNA caused by an agent or mutagen. Mutagens often demonstrate mutational specificity, meaning they cause predictable changes in the DNA sequence.[5] A few common mutagens that induce mutations are:
- Ultraviolet light (UV) – Causes pyrimidine (T or C) nucleotide bases on the same strand to covalent join forming a pyrimidine dimer. Thymine-thymine dimers are the most common mutation caused by UV light. Since dimers cause a disruptive kink in DNA structure, polymerases often have trouble reading the region, slowing down DNA replication.[5]
- Base analogs – Chemical compounds that are sufficiently similar in structure and chemistry to the nitrogenous bases of DNA, such that they are able to be incorporated in the sequence. These analogs do not have the same pairing properties of normal bases, therefore they can pair incorrectly with nucleotides during replication. 5-bromouracil (5-BU) is a common analog to thymine, however the enol form of 5-BU is still able to bind with adenine. The ionized form, on the other hand, pairs with guanine.[5]
- Intercalating agents – Chemical compounds that place themselves between stacked nitrogenous bases in DNA, causing a frameshift mutation. Some intercalating agents, like daunorubicin, are capable of blocking replication and transcription, making them incredibly toxic to proliferating cells.[5]
- Reactive oxygen species (ROS) – Highly reactive oxygen-containing molecules that are capable causing DNA strand breaks and many damaging effects to cellular components.[5][6]
- Alkylating agents – Compounds that attach an alkyl group to the four bases. When an alkyl group is added to guanine, it can lead to the incorrect pairing with thymine and disrupt the accuracy of replication.[5]
Consequences
A large determinant of the severity of consequences caused by postzygotic mutations is when and where they occur. As a result, consequences can range from being negligible to incredibly detrimental.[7]
Mosaicism
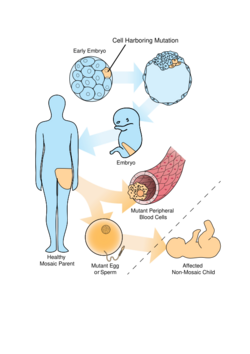
When an individual has inherited an abnormality it is usually present in all of their cells. However some mutations like DNA code change, epigenetic alterations and chromosomal abnormalities, can occur later in development. This would result in one progeny cell line to be normal while the other cell line(s) to be abnormal. As a result, the individual is considered to be a mosaic of normal and abnormal cells.[7]
Mosaicism is the occurrence of two or more cell lines with different genotypes within a single individual. It is different from chimerism which is the fusion of two zygotes, causing a new single zygote with two genotypes.[7]
Loss of chromosome Y
The loss of chromosome Y (LOY) in blood cells is the most common human postzygotic mutation. It is highly associated with age, being detectable in at least 10% of blood cells for 14% and 57% of males around 70 and 94 years of age, respectively.[8][9] Men with LOY have a higher all-cause mortality and cancer mortality compared with unaffected males.[10] Additionally, LOY is associated with greater risk for Alzheimer's disease and cardiovascular disease.[11] Smoking increases the risk of inducing LOY more than three times and has a dose-dependent effect on LOY-status.[12]
Trisomy 21 mosaicism
Trisomy 21 (Down syndrome) is one of the most prevalent chromosomal abnormalities amongst live births. Of all trisomy 21 pregnancies, approximately 80% end in spontaneous abortions or still-births. 1–5% of people diagnosed with having Down Syndrome are actually in fact "high-grade" trisomy 21 mosaics. The rest of trisomy 21 mosaics are marked as "low-grade" mosaics, meaning the chromosomal mutation occurs in less than 3–5% of respective tissue. While high-grade trisomy 21 mosaics, demonstrate similar features to full Down Syndrome, low-grade mosaics have a tendency to show milder features; however, the effects are quite variable depending on the distribution of the trisomic cells.[13]
Somatic mutations
Somatic mutations are the result of a change in the genetic structure after fertilization. This type of mutation also involves cells outside of the reproductive group and thus is not transmitted to future descendants.[14][15]
Germ-line mutations
Germ-line mutations are the result of a change in the genetic structure of germ cells. These mutations are able to be transmitted to the offspring and give rise to a constitutional mutation. Constitutional mutations is a mutation that when present in one cell, is also present in all other cells associated with the organism.[15]
References
- ↑ Acuna-Hidalgo, Rocio; Bo, Tan; Kwint, Michael P.; van de Vorst, Maartje; Pinelli, Michele; Veltman, Joris A.; Hoischen, Alexander; Vissers, Lisenka E. L. M. et al. (2015-07-02). "Post-zygotic Point Mutations Are an Underrecognized Source of De Novo Genomic Variation". The American Journal of Human Genetics 97 (1): 67–74. doi:10.1016/j.ajhg.2015.05.008. PMID 26054435.
- ↑ 2.0 2.1 2.2 2.3 2.4 Griffiths, Anthony JF; Miller, Jeffrey H.; Suzuki, David T.; Lewontin, Richard C.; Gelbart, William M. (2000-01-01). "Spontaneous mutations". An Introduction to Genetic Analysis. https://www.ncbi.nlm.nih.gov/books/NBK21897/.[|permanent dead link|dead link}}]
- ↑ 3.0 3.1 Alberts, Bruce; Johnson, Alexander; Lewis, Julian; Raff, Martin; Roberts, Keith; Walter, Peter (2002-01-01). "DNA Repair". Molecular Biology of the Cell (4th ed.). https://www.ncbi.nlm.nih.gov/books/NBK26879/.
- ↑ "Documents – All Documents". http://faculty.ksu.edu.sa/77379/Documents/. Retrieved 2015-12-02.
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 Griffiths, Anthony JF; Miller, Jeffrey H.; Suzuki, David T.; Lewontin, Richard C.; Gelbart, William M. (2000-01-01). "Induced mutations". An Introduction to Genetic Analysis. https://www.ncbi.nlm.nih.gov/books/NBK21936/.[|permanent dead link|dead link}}]
- ↑ Cooke, Marcus S.; Evans, Mark D.; Dizdaroglu, Miral; Lunec, Joseph (2003-07-01). "Oxidative DNA damage: mechanisms, mutation, and disease". The FASEB Journal 17 (10): 1195–1214. doi:10.1096/fj.02-0752rev. ISSN 0892-6638. PMID 12832285.
- ↑ 7.0 7.1 7.2 Youssoufian, Hagop; Pyeritz, Reed E. (2002-10-01). "Mechanisms and consequences of somatic mosaicism in humans". Nature Reviews. Genetics 3 (10): 748–758. doi:10.1038/nrg906. ISSN 1471-0056. PMID 12360233.
- ↑ Forsberg, Lars A. (May 2017). "Loss of chromosome Y (LOY) in blood cells is associated with increased risk for disease and mortality in aging men" (in en). Human Genetics 136 (5): 657–663. doi:10.1007/s00439-017-1799-2. ISSN 0340-6717. PMID 28424864.
- ↑ Forsberg, Lars A.; Halvardson, Jonatan; Rychlicka-Buniowska, Edyta; Danielsson, Marcus; Moghadam, Behrooz Torabi; Mattisson, Jonas; Rasi, Chiara; Davies, Hanna et al. (January 2019). "Mosaic loss of chromosome Y in leukocytes matters" (in en). Nature Genetics 51 (1): 4–7. doi:10.1038/s41588-018-0267-9. ISSN 1546-1718. PMID 30374072.
- ↑ Forsberg, Lars A; Rasi, Chiara; Malmqvist, Niklas; Davies, Hanna; Pasupulati, Saichand; Pakalapati, Geeta; Sandgren, Johanna; Ståhl, Teresita Diaz de et al. (June 2014). "Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer". Nature Genetics 46 (6): 624–628. doi:10.1038/ng.2966. ISSN 1546-1718. PMID 24777449.
- ↑ Haitjema, Saskia; Kofink, Daniel; Setten, Jessica van; Laan, Sander W. van der; Schoneveld, Arjan H.; Eales, James; Tomaszewski, Maciej; Jager, Saskia C. A. de et al. (2017-08-01). "Loss of y Chromosome in Blood is Associated with Major Cardiovascular Events During Follow-Up in Men After Carotid Endarterectomy". Circulation: Cardiovascular Genetics 10 (4): e001544. doi:10.1161/circgenetics.116.001544. ISSN 1942-325X. PMID 28768751.
- ↑ Dumanski, Jan P.; Rasi, Chiara; Lönn, Mikael; Davies, Hanna; Ingelsson, Martin; Giedraitis, Vilmantas; Lannfelt, Lars; Magnusson, Patrik K. E. et al. (2015-01-02). "Smoking is associated with mosaic loss of chromosome Y". Science 347 (6217): 81–83. doi:10.1126/science.1262092. ISSN 0036-8075. PMID 25477213. Bibcode: 2015Sci...347...81D.
- ↑ Hultén, Maj A.; Jonasson, Jon; Nordgren, Ann; Iwarsson, Erik (2010-09-01). "Germinal and Somatic Trisomy 21 Mosaicism: How Common is it, What are the Implications for Individual Carriers and How Does it Come About?". Current Genomics 11 (6): 409–419. doi:10.2174/138920210793176056. ISSN 1389-2029. PMID 21358985.
- ↑ "Somatic mutation – Glossary Entry". 2015-11-09. http://ghr.nlm.nih.gov/glossary=somaticmutation. Retrieved 2015-11-14.
- ↑ 15.0 15.1 Griffiths, Anthony JF; Miller, Jeffrey H.; Suzuki, David T.; Lewontin, Richard C.; Gelbart, William M. (2000-01-01). "Somatic versus germinal mutation". An Introduction to Genetic Analysis. https://www.ncbi.nlm.nih.gov/books/NBK21894/.[|permanent dead link|dead link}}]
 |
