Biology:PyClone
PyClone is a software that implements a Hierarchical Bayes statistical model to estimate cellular frequency patterns of mutations in a population of cancer cells using observed alternate allele frequencies, copy number, and loss of heterozygosity (LOH) information. PyClone outputs clusters of variants based on calculated cellular frequencies of mutations.[1]
Background

According to the Clonal Evolution model proposed by Peter Nowell, a mutated cancer cell can accumulate more mutations as it progresses to create sub-clones. These cells divide and mutate further to give rise to other sub-populations. In compliance with the theory of natural selection, some mutations may be advantageous to the cancer cells and thus make the cell immune to previous treatment. Heterogeneity within a single cancer tumour can arise from single nucleotide polymorphism/variation (SNP/SNV) events, microsatellite shifts and instability, loss of heterozygosity (LOH), Copy number variation and karyotypic variations including chromosome structural aberrations and aneuploidy. Due to the current methods of molecular analysis where a mixed population of cancer cells are lysed and sequenced, heterogeneity within the tumour cell population is under-detected. This results in a lack of information on the clonal composition of cancer tumours and more knowledge in this area would aid in the decisions for therapies.
PyClone is a hierarchical Bayes statistical model that uses measurements of allele frequency and allele specific copy numbers to estimate the proportion of tumor cells harboring a mutation. By using deeply sequenced data to find putative clonal clusters, PyClone estimates the cellular prevalence, the portion of cancer cells harbouring a mutation, of the input sample. Progress has been made for measuring variant allele frequency with deep sequencing data but statistical approaches to cluster mutations into biologically relevant groups remain underdeveloped. The commonness of a mutation between cells is difficult to measure because the proportion of cells that harbour a mutation doesn't simply relate to allelic prevalence. This is due to allelic prevalence depending on multiple factors such as the proportion of 'contaminating' normal cells in the sample, the proportion of tumor cells harboring the mutation, the number of allelic copies of the mutation in each cell, and sources of technical noise. PyClone is among the first methods to incorporate variant allele frequencies (VAFs) with allele-specific copy numbers.[2] It also accounts for Allelic Imbalances, where alleles of a gene are expressed at different levels in a given cell,[3] which may occur in the cell due to Segmental CNVs and normal cell contamination.
Workflow
Input

PyClone requires 2 inputs:
- A set of deeply sequenced mutations from one or more samples derived from a single patient. Deep sequencing, also referred to as high throughput sequencing, uses methods such as sequencing by synthesis to sequence a genomic region with high coverage in order to detect rare clonal types and contaminating normal cells that comprise as little as 1% of the sample.[4]
- A measure of allele specific copy number at each mutation location. This is obtained from microarray-based comparative genomic hybridization or whole genome sequencing methods to detect chromosomal or copy number changes.
Statistical modeling
For each mutation, the PyClone model divides the input sample into three sub-populations. The three sub-populations are the normal (non-malignant) population consisting of normal cells, the reference cancer population consisting of cancer cells wild type for the mutation, and the variant cancer cell population consisting of the cancer cells with at least one variant allele of the mutation.
PyClone implements four advances in its statistic model that were tested on simulated datasets :
Beta-binomial emission densities
Beta-binomial Emission Densities are used by PyClone and are more effective than binomial models used by previous tools. Beta-binomial emission densities more accurately model input datasets that have more variance in allelic prevalence measurements. Higher accuracy in modeling variance in allelic prevalence translates to a higher confidence in the clusterings outputted by PyClone.
Priors
PyClone acknowledges that some geometrical structures and properties, such as copy number, of the clonal population to be reconstructed is known. When not enough information is available or taken into account, the reconstruction is usually of low confidence and many solutions are possible. PyClone uses priors, flexible prior probability estimates, of possible mutational genotypes to link allelic prevalence measurements to zygosity and copy number variants and is one of the first methods to incorporate variant allele frequencies (VAFs) with allele-specific copy numbers.[5]
Bayesian nonparametric clustering
Instead of fixing the number of clusters prior to clustering, Bayesian nonparametric clustering is used to discover groupings of mutations and the number of groups simultaneously. This allows for cellular prevalence estimates to reflect uncertainty in this parameter.
Section sequencing
Multiple samples from the same patient can be analyzed at the same time to leverage the scenario in which clonal populations are shared across samples. When multiple samples are sequenced, subclonal populations that are similar in allelic prevalence in some cells but not others can be differentiated from each other.
Output
PyClone outputs posterior densities of cellular prevalences for the mutations in the sample and a matrix containing the probability any two mutations occur in the same cluster. Estimates of clonal populations from differing cellular prevalences of mutations are then generated from the posterior densities.
Applications
PyClone is used to analyze deeply sequenced (over 100× coverage) mutations to identify and quantify clonal populations in tumors. Some applications include:
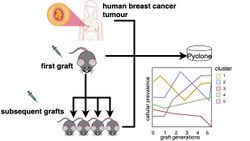
Xenografting is used as a reasonable model to study human breast cancer but the consequences of engraftment and genomic propagation of xenografts have not been examined at a single-cell resolution. PyClone can be used to follow the clonal dynamics of initial grafts and serial propagation of primary and metastatic human breast cancers in immunodeficient mice. PyClone can predict how clonal dynamics differ after initial engraftment, over serial passage generations.[6]
Circulating tumour DNA (plasma DNA) Analysis can be used to track tumour burden and analyse cancer genomes non-invasively but the extent to which it represents metastatic heterogeneity is unknown. PyClone can be used to compare the clonal population structures present in the tumour and plasma samples from amplicon sequencing data. Stem and metastatic-clade mutation clusters can be inferred using PyClone and then compared to results from clonal ordering.[7]
Serial Time Point Sequencing: PyClone can be used to study the evolution of mutational clusters as cancer progresses. With samples taken from different time points, PyClone can identify the expansion and decline of initial clones and discover newly acquired subclones that arise during treatment. Understanding clonal dynamics improves understanding on how related cancers such as MDS, MPN and sAML compare in risk and give insight on the clinical significance of somatic mutations.[8]
Section sequencing: PyClone is most effective for section sequencing tumor DNA. Section sequencing is when samples are taken from different portions of a single tumour to infer clonal structure from differential cellular prevalence. An advantage of section sequencing is more statistical power and information on the spatial position and interactions of the clones, uncovering information on how tumors evolve in space.
Assumptions
A key assumption of the PyClone model is that all cells within a clonal population have the same genotype. This assumption is likely false since copy number alterations and loss of heterozygosity events are common in cancer cells. The amount of error introduced by this assumption depends on the variability of genotype of cells in the location of interest. For example, in solid tumors the cells of a sample are spatially close together resulting in a small error rate, but for liquid tumors the assumption may introduce more error as cancer cells are mobile.
Another assumption made is that the sample follows a perfect and persistent phylogeny. This means that no site mutates more than once in a clonal population and each site has at most one mutant genotype. Mutations that revert to normal genotype, deletions of segments of DNA harbouring mutations and recurrent mutations are not accounted for in PyClone as it would lead to unidentifiable explanations for some observed data.
Limitations
In order to obtain input data for PyClone, cell lysis is a required step to prepare bulk sample sequencing. This results in the loss of information on the complete set of mutations defining a clonal population. PyClone can distinguish and identify the frequency of different clonal populations but can not identify exact mutations defining these populations.
Instead of clustering cells by mutational composition, PyClone clusters mutations that have similar cellular frequencies. In sub-clones that have similar cellular frequencies, PyClone will mistakenly cluster these subclones together. Chances of making this error decreases when using targeted deep sequencing with high coverage and joint analysis of multiple samples
A confounding factor of the PyClone model arises due to imprecise input information on the genotype of the sample and the depth of sequencing. Uncertainty arises in the posterior densities due to insufficient information on the genotype of mutations and depth of sequencing of the sample. This results in relying on the assumptions made by the PyClone model to interpret and cluster the sample.
Similar tools
SciClone[9]- SciClone is a Bayesian clustering method on single nucleotide variants (SNVs).
Clomial[10]- Clomial is a Bayesian clustering method with a decomposition process. Both Clomial and SciCloe limit the SNVs located in copy-number neutral region. The tumor is physically divided into subsections and deep sequenced to measure normal allele and variant allele. Their inference model uses Expectation-Maximization algorithm.
GLClone[11] – GLClone uses a hierarchical probabilistic model and Bayesian posteriors to calculate copy number alterations in sub-clones.
Cloe [12]- Cloe uses a phylogenetic latent feature model for analyzing sequencing data to distinguish the genotypes and the frequency of clones in a tumor.
PhyC [13]- PhyC uses an unsupervised learning approach to identify subgroups of patients through clustering the respective cancer evolutionary trees. They identified the patterns of different evolutionary modes in a simulation analysis, and also successfully detected the phenotype-related and cancer type-related subgroups to characterize tree structures within subgroups using actual datasets.
PhyloWGS [14]- PhyloWGS reconstructs tumor phylogenies and characterizes the subclonal populations present in a tumor sample using both SSMs and CNVs.
References
- ↑ Fearon, Eric R.; Wu, Rong; Greenson, Joel K.; Ulintz, Peter J.; Hardiman, Karin M. (2017-02-01). "Abstract PR07: Complex sub-clonal populations in colorectal cancer lymph node metastasis". Cancer Research 77 (3 Supplement): PR07. doi:10.1158/1538-7445.CRC16-PR07. ISSN 0008-5472. http://cancerres.aacrjournals.org/content/77/3_Supplement/PR07.
- ↑ 2.0 2.1 Shah, Sohrab P.; Bouchard-Côté, Alexandre; Aparicio, Samuel; Ha, Gavin; Justina Biele; Laks, Emma; Wan, Adrian; Yap, Damian et al. (April 2014). "PyClone: statistical inference of clonal population structure in cancer". Nature Methods 11 (4): 396–398. doi:10.1038/nmeth.2883. ISSN 1548-7105. PMID 24633410.
- ↑ Blanchette, Mathieu; Pastinen, Tomi; Gunderson, Kevin L.; Pokholok, Dmitry; Ge, Bing; Wagner, James R. (2010-07-08). "Computational Analysis of Whole-Genome Differential Allelic Expression Data in Human". PLOS Computational Biology 6 (7): e1000849. doi:10.1371/journal.pcbi.1000849. ISSN 1553-7358. PMID 20628616.
- ↑ "Deep Sequencing". https://www.illumina.com/science/technology/next-generation-sequencing/deep-sequencing.html.
- ↑ Hanson, K.M. (May 1993). "Bayesian reconstruction based on flexible prior models". J. Opt. Soc. Am. A 10 (5): 997. doi:10.1364/JOSAA.10.000997.
- ↑ Aparicio, Samuel; Shah, Sohrab P.; Caldas, Carlos; Marra, Marco A.; Hansen, Carl; Eaves, Connie J.; Huntsman, David; Nguyen, Long et al. (February 2015). "Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution". Nature 518 (7539): 422–426. doi:10.1038/nature13952. ISSN 1476-4687. PMID 25470049.
- ↑ Caldas, Carlos; Rosenfeld, Nitzan; Wallis, Matthew; Shah, Sohrab P.; Bentley, David; Humphray, Sean; Kingsbury, Zoya; Shumansky, Karey et al. (2015-11-04). "Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer". Nature Communications 6: 8760. doi:10.1038/ncomms9760. ISSN 2041-1723. PMID 26530965.
- ↑ Maciejewski, Jaroslaw P.; Ogawa, Seishi; Shih, Lee-Yung; Miyano, Satoru; Chiba, Shigeru; Saunthararajah, Yogen; Miyawaki, Shuichi; Nakamaki, Tsuyoshi et al. (2015-12-03). "Serial Sequencing in Myelodysplastic Syndromes Reveals Dynamic Changes in Clonal Architecture and Allows for a New Prognostic Assessment of Mutations Detected in Cross-Sectional Testing" (in en). Blood 126 (23): 709. doi:10.1182/blood.V126.23.709.709. ISSN 0006-4971. http://www.bloodjournal.org/content/126/23/709.
- ↑ Ding, Li; Wilson, Richard K.; Mardis, Elaine R.; Ley, Timothy J.; DiPersio, John F.; Schierding, William; Ellis, Matthew J.; Walter, Matthew J. et al. (2014-08-07). "SciClone: Inferring Clonal Architecture and Tracking the Spatial and Temporal Patterns of Tumor Evolution" (in en). PLOS Computational Biology 10 (8): e1003665. doi:10.1371/journal.pcbi.1003665. ISSN 1553-7358. PMID 25102416.
- ↑ Noble, William Stafford; Blau, C. Anthony; Witten, Daniela; Song, ChaoZhong; Nickerson, Debbie; Smith, Josh; Weber, Kris; Hu, Alex et al. (2014-07-10). "Inferring Clonal Composition from Multiple Sections of a Breast Cancer" (in en). PLOS Computational Biology 10 (7): e1003703. doi:10.1371/journal.pcbi.1003703. ISSN 1553-7358. PMID 25010360.
- ↑ Geng, Yu; Zhao, Zhongmeng; Xu, Jing; Liu, Ruoyu; Huang, Yi; Zhang, Xuanping; Xiao, Xiao; Maomao et al. (2017). "Identifying Heterogeneity Patterns of Allelic Imbalance on Germline Variants to Infer Clonal Architecture". in Huang, De-Shuang; Jo, Kang-Hyun; Figueroa-García, Juan Carlos (in en). Intelligent Computing Theories and Application. Lecture Notes in Computer Science. 10362. Springer International Publishing. pp. 286–297. doi:10.1007/978-3-319-63312-1_26. ISBN 978-3-319-63312-1.
- ↑ Markowetz, Florian; Rosenfeld, Nitzan; Yuan, Ke; Mouliere, Florent; Marass, Francesco (2016-04-06). "A phylogenetic latent feature model for clonal deconvolution" (in en). The Annals of Applied Statistics 10 (4): 2377–2404. doi:10.1214/16-AOAS986.
- ↑ Shimamura, Teppei; Miyano, Satoru; Mimori, Koshi; Uchi, Ryutaro; Niida, Atsushi; Matsui, Yusuke (2017-05-01). "phyC: Clustering cancer evolutionary trees" (in en). PLOS Computational Biology 13 (5): e1005509. doi:10.1371/journal.pcbi.1005509. ISSN 1553-7358. PMID 28459850.
- ↑ Deshwar, Amit G.; Vembu, Shankar; Yung, Christina K.; Jang, Gun Ho; Stein, Lincoln; Morris, Quaid (2015-02-13). "PhyloWGS: Reconstructing subclonal composition and evolution from whole-genome sequencing of tumors". Genome Biology 16 (1): 35. doi:10.1186/s13059-015-0602-8. ISSN 1465-6906. PMID 25786235.
 |
