Biology:S1 domain
| S1 domain | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
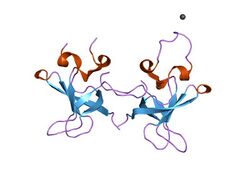 Crystal structure of the S1 domain of RNase E from E. coli (Pb derivative).[1] | |||||||||||
| Identifiers | |||||||||||
| Symbol | S1 | ||||||||||
| Pfam | PF00575 | ||||||||||
| Pfam clan | CL0021 | ||||||||||
| InterPro | IPR003029 | ||||||||||
| SMART | S1 | ||||||||||
| PROSITE | PDOC00053 | ||||||||||
| MEROPS | S15 | ||||||||||
| SCOP2 | 1sro / SCOPe / SUPFAM | ||||||||||
| CDD | cd00164 | ||||||||||
| |||||||||||
The S1 domain is a protein domain that was originally identified in ribosomal protein S1 but is found in a large number of RNA-associated proteins. The structure of the S1 RNA-binding domain from the Escherichia coli polynucleotide phosphorylase has been determined using NMR methods and consists of a five-stranded antiparallel beta barrel. Conserved residues on one face of the barrel and adjacent loops form the putative RNA-binding site.[2]
The structure of the S1 domain is very similar to that of cold shock proteins. This suggests that they may both be derived from an ancient nucleic acid-binding protein.[2]
Function
The S1 domain is an essential in protein translation as it interacts with the ribosome and messenger RNA. S1 bind to RNA in a sequence specific manner.
Structure
This protein domain contains six motifs and 70 amino acids and it folds into a five-stranded antiparallel beta barrel. The structure of the S1 domain is very similar to that of cold shock proteins. This suggests that they may both be derived from an ancient nucleic acid-binding protein. Conserved residues on one face of the barrel and adjacent loops form the putative RNA-binding site.[2]
References
- ↑ Schubert M; Edge RE; Lario P et al. (July 2004). "Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces". J. Mol. Biol. 341 (1): 37–54. doi:10.1016/j.jmb.2004.05.061. PMID 15312761.
- ↑ Jump up to: 2.0 2.1 2.2 "The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold". Cell 88 (2): 235–42. January 1997. doi:10.1016/S0092-8674(00)81844-9. PMID 9008164.
 |

