Chemistry:Klebsazolicin
| |
| Names | |
|---|---|
| Other names
KLB
| |
| Identifiers | |
3D model (JSmol)
|
|
PubChem CID
|
|
| |
| |
| Properties | |
| C76H107N27O30S3 | |
| Molar mass | 1975.03 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
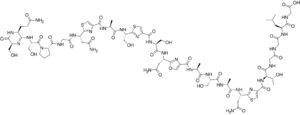
Klebsazolicin (KLB) is a peptide antibiotic encoded in the genome of a gram-negative bacterium Klebsiella pneumoniae subsp. ozonae and targeting prokaryotic ribosome.[1][2] Klebsazolicin is a ribosomally synthesized and post-translationally modified peptide (RiPP) and a linear azol(in)e-containing peptide (LAP).
Discovery
The discovery of KLB represents an example of a “genome mining” approach. Cluster of genes encoding KLB biosynthetic pathway was found in genome database using low-level homology of one of the proteins to microcin B17 synthetase. Cloning and expression of cluster in a heterologous host (Escherichia coli) yielded the active compound. The name given to the compound reflects the original bacterium where the biosynthetic cluster was found (Klebs-) and the presence of azole cycles (-azolicin).[citation needed]
Structure
The structure of KLB was established by a combination of mass-spectrometry and NMR methods. Klebsazolicin is 23-aminoacid long peptide, containing four azoles (3 thiazoles and an oxazole) and an N-terminal lactamidine ring. The latter is formed by a linkage between two N-terminal amino acids (serine and glutamine) and is absolutely essential for bioactivity.[3]
Activity
Klebsazolicin is active against Gram-negative bacteria closely related to Klebsiella, such as Escherichia coli, Klebsiella pneumoniae, and Yersinia pseudotuberculosis. KLB inhibits protein synthesis on the prokaryotic ribosome by binding to and blocking peptide exit tunnel and thus preventing the passage of the nascent peptide. The activity of KlpE export pump encoded in KLB biosynthetic gene cluster confers self-resistance of the producing bacterium to the action of the antibiotic.[citation needed]
Biosynthesis
Typically to other RiPPs, klebsazolicin is produced in three steps. At the first step, a 47-aa precursor peptide KlpA is synthesized using cellular translation machinery. Then an N-terminal leader peptide serves as a recognition element for KlpBCD, a heterocyclase-dehydrogenase complex which converts serine and cysteine residues of KlpA into oxazole and thiazole heterocycles. Finally, the leader is cleaved off by the action of cellular proteases such as TldD/E, and at the same time KlpBCD activates the new N-terminus to form lactamidine. Thus, KlpBCD is able to introduce both azole heterocycles and lactamidine linkages, using side chains of Ser/Cys residues and N-terminal amino group as nucleophiles.[4]
References
- ↑ "Structure Summary for 5W4K". U.S.: Rutgers University. http://ndbserver.rutgers.edu/service/ndb/atlas/summary?searchTarget=5W4K.
- ↑ Bank, RCSB Protein Data. "RCSB PDB - PRD_002277 BIRD Summary Page". Protein Data Bank. https://www.rcsb.org/bird/PRD_002277.
- ↑ Metelev, M; Osterman, IA; Ghilarov, D; Khabibullina, NF; Yakimov, A; Shabalin, K; Utkina, I; Travin, DY et al. (October 2017). "Klebsazolicin inhibits 70S ribosome by obstructing the peptide exit tunnel.". Nature Chemical Biology 13 (10): 1129–1136. doi:10.1038/nchembio.2462. PMID 28846667.
- ↑ Travin, DY; Metelev, M; Serebryakova, M; Komarova, ES; Osterman, IA; Ghilarov, D; Severinov, K (25 April 2018). "Biosynthesis of Translation Inhibitor Klebsazolicin Proceeds through Heterocyclization and N-Terminal Amidine Formation Catalyzed by a Single YcaO Enzyme.". Journal of the American Chemical Society 140 (16): 5625–5633. doi:10.1021/jacs.8b02277. PMID 29601195.
 |

