Chemistry:Moenomycin family antibiotics
First described in 1965, the moenomycins are a family of phosphoglycolipid antibiotics, metabolites of the bacterial genus Streptomyces. Moenomycin A is the founding member of the antibiotic family with the majority discovered by the end of the late 1970s.[1]
Structure
The moenomycins can be reduced to three key structural features[2]
- A central 3-phosphoglyceric acid backbone.
- A 25-carbon isoprenoid chain connected by an ether linkage to the C2-position of 3-phosphoglyceric acid.
- A substituted tetrasaccharide tethered via a phosphodiester linkage to 3-phosphoglyceric acid.
It is the combination of different isoprenoid chains and variously substituted tetrasaccharides that give rise to the diversity of the moenomycin family.[3]
Based on degradation experiments, the defining mark of a moenomycin is the presence of the 25-carbon alcohol moenocinol or diumycinol upon hydrolysis of the lipid tail; these alcohols originate from the L1 or L2 lipid respectively in the figure.[4] These two structures are the only observed lipid tails within the moenomycin family, with AC326-α being the only known for producing diumycinol.[5]
With regards to the tetrasaccharide portion, stereochemistry and functionality can differ at R1 and R2 depending on if this saccharide unit is D-gluco versus D-galacto; there is an axial methyl group in the former case with the exception of moenomycin A12 and C1 where there is instead an axial hydroxyl. The oligosaccharide motif can be deoxygenated, hydroxylated, or glycosylated at the R3 position – notable examples of the pentasaccharide motif include moenomycin A and AC326-α. It is believed the additional glycan can enhance specificity and binding to the target protein, affording increased activity.[6] With the exception of pholipomycin and AC326-α, the R4 saccharide unit is usually the deoxysaccharide. Lastly, in the majority of moenomycins the R5 position is linked to a 2-aminocyclopentane-1,3-dione – a convenient chromophore utilized for structural analysis. For the nosokomycin subfamily, this position forms a carboxamide or carboxylic acid.[7][8]
Chemical synthesis
Due to the structural complexity of the moenomycins, total synthesis has proved difficult, with only one total synthesis reported so far. Some of the largest challenges include fashioning the glycosidic linkages with stereochemical control and site-specifically decorating the oligosaccharide with pendant functionality. Understanding that the majority of variation within the moenomycin family derives from differences within the oligosaccharide unit, Kahne and lab has designed an efficient and flexible total synthesis of moenomycin A that gives access to analogues as well as other members of the moenomycin family.[9]
Biosynthesis
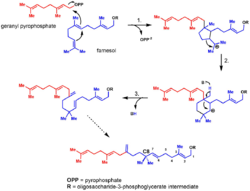
Extensive exploration into the biosynthesis of the moenomycin family has been conducted to better inform the genetic engineering and biosynthesis of novel moenomycin analogues. Early work on the biosynthesis of the moenomycins focused on the 25-carbon lipid tail derived from moenocinol; the tail was of particular interest given that it appears to break the isoprene rule at C8, containing a quaternary carbon. Feeding studies revealed the moenocinol lipid tail originates from a 15-carbon farnesyl precursor and a 10-carbon geranyl pyrophosphate.[10]
More recently, the biosynthetic gene cluster for moenomycin A was first described in 2007 in Streptomyces ghanaensis.[11] In 2009, the seventeen step biosynthetic pathway was completely characterized, revealing the order of assembly for the molecular scaffold.[12]
Medicinal use
The moenomycins target bacterial peptidoglycan glycosyltransferases, inhibiting cell wall formation, leading to cell death.[13] In general, the antibiotics are particularly potent against gram-positive bacteria with a minimum inhibitory concentration (MIC) between 1–100 (ng/ml). At higher concentrations the moenomycins are also effective against gram-negative bacteria with an MIC between 0.3–150 (μg/ml). In vivo studies using mice models suggest the antibiotics are powerful prophylactic and therapeutic agents, with subcutaneous injection being the most effective mode of delivery.[14]
Moenomycins A and C are commercially used in the formulation of Bambermycins (Flavomycin), a veterinary antibiotic used solely in poultry, swine, and cattle feed.[15]
Due to poor pharmacokinetic properties from the 25-carbon lipid chain, the moenomycins are not used in humans. The pharmacophore is well understood however, allowing the moenomycins to serve as the blueprint for future antibacterials.[7]
Mode of action
General
The moenomycin family functions as an antibiotic by reversibly binding bacterial transglycosylases, essential enzymes that catalyze the extension of the glycan chain of the cell wall to form a stable peptidoglycan layer. The moenomycins mimic and thus compete with the natural substrate of the enzyme, inhibiting growth of the cell wall. Compromise of the wall results in leakage of cell contents, and ultimately cell death. The moenomycins are the only known active site inhibitors of these enzymes, which in lies their promise as human antibiotics given pathogenic bacteria have not yet widely evolved resistance.[16]
Structure-activity relationships
The 25-carbon lipid tail confers to the moenomycins a detergent-like property that allows them to become incorporated into the cytoplasmic membrane of the target bacterial cell. This anchoring presents the oligosaccharide portion of the molecule to the transglycosylase where it can tightly and selectively bind the enzyme, inhibiting cell wall growth.[17] This property however undermines their use in clinical settings. The amphiphilic nature of the moenomycins induce hemolytic activity, provide a long half-life in the blood stream, and creates a tendency to aggregate in aqueous solution. Comparison of moenomycins with an abridged isoprene chain of 10-carbons, show that the oligosaccharide can still tightly bind the enzyme active site, but in vivo the MIC significantly increases since the drug is unable to anchor itself to the cytoplasmic membrane and present its sugar moiety. Further studies are needed to determine the optimal length for favorable pharmacokinetic properties.[18]
In contrast to the lipid portion, the oligosaccharide portion of the moenomycins is relatively well understood. When absent, the chromophore portion can decrease activity by 10-fold, suggesting it is not necessary for recognition but provides additional contacts with the target enzyme.[19]
References
- ↑ Wallhausser, K.H., et al. Antimicrob Agents Chemother. 1965. P. 734-736.
- ↑ Walker, S., Ostash, B.O., Moenomycin Biosynthesis-related Compositions and Methods of Use Thereof . U.S. Patent 9115358 B2, Aug 25, 2015.
- ↑ Welzel, P. Chem. Rev. 2005, 105, 4610−4660
- ↑ Adachi, M., et al. J. Am. Chem. Soc., 2006, 128 (43), pp 14012–14013
- ↑ HE, HAIYIN; SHEN, BO; KORSHALLA, JOSEPH; SIEGEL, MARSHALL M.; CARTER, GUY T. (25 February 2000). "Isolation and Structural Elucidation of AC326-.ALPHA., A New Member of the Moenomycin Group.". The Journal of Antibiotics 53 (2): 191–195. doi:10.7164/antibiotics.53.191. PMID 10805581. https://www.jstage.jst.go.jp/article/antibiotics1968/53/2/53_2_191/_article.
- ↑ Yuan, Y. ACS Chem. Biol., 2008, 3 (7), pp 429–436
- ↑ 7.0 7.1 Ostash, B.; Walker, S. Nat. Prod. Rep., 2010, 27, 1594-1617
- ↑ Schacht U, Huber G J Antibiot (Tokyo). 1969 Dec; 22(12):597-602.
- ↑ Kahne, D.E., et al. J. Am. Chem. Soc., 2006, 128 (47), pp 15084–15085
- ↑ Arigoni, D., et al. Tetrahedron Letters, 2001, 42, 3835–37
- ↑ Ostash B., et al. Chem Biol. 2007, 14, 257-67.
- ↑ Ostash, B., et al. Biochemistry 2009, 48, 8830–41.
- ↑ Gampe, C. J. Am. Chem. Soc., 2013, 135 (10), pp 3776–3779
- ↑ Huber G. In: Antibiotics. Hahn FE, editor. Springer-Verlag; Berlin/Heidelberg: 1979. pp. 135–153.
- ↑ National Center for Biotechnology Information. PubChem Compound Database; CID=53385491, https://pubchem.ncbi.nlm.nih.gov/compound/53385491 (accessed Mar. 8, 2017).
- ↑ Walsh, C.T., The Journal of Antibiotics (2014) 67, 7–22
- ↑ Volke, F. Chemistry and Physics of Lipids 85 (1997) 115–123.
- ↑ Anikin, A., et al. Angew Chem Int Ed Engl. 1999 Dec 16; 38(24):3703-3707.
- ↑ T. Ru¨hl et al. / Bioorg. Med. Chem. 11 (2003) 2965–2981
 |