Chemistry:Pyoluteorin
 | |
| Identifiers | |
|---|---|
| |
| CAS Number | |
| PubChem CID | |
| ChemSpider | |
| UNII | |
| ChEBI | |
| ChEMBL | |
| Chemical and physical data | |
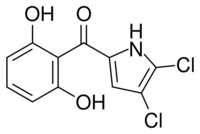
| Formula | C11H7Cl2NO3 |
| Molar mass | 272.08 g·mol−1 |
| 3D model (JSmol) | |
| |
| |
Pyoluteorin is a natural antibiotic that is biosynthesized from a hybrid nonribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) pathway.[1] Pyoluteorin was first isolated in the 1950s from Pseudomonas aeruginosa strains T359 and IFO 3455[2] and was found to be toxic against oomycetes, bacteria, fungi, and against certain plants.[3] Pyoluteorin is most notable for its toxicity against the oomycete Pythium ultimum,[4] which is a plant pathogen that causes a global loss in agriculture. Currently, pyoluteorin derivatives are being studied as an Mcl-1 antagonist in order to target cancers that have elevated Mcl-1 levels.[5]
Biosynthesis
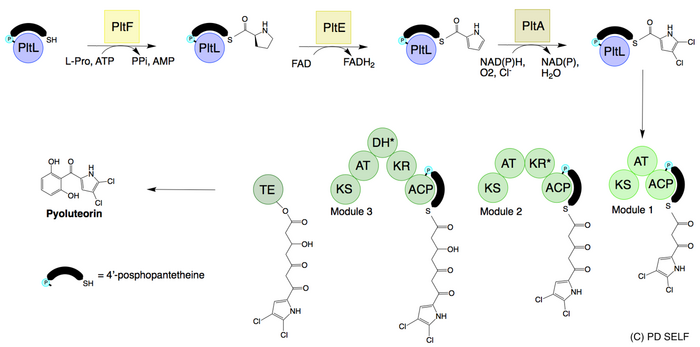
Pyoluteorin is synthesized from an NRPS/PKS hybrid pathway. The resorcinol ring is derived from a type I PKS[6][7] while the dichloropyrrole[clarification needed] moiety is derived from a type II NRPS.[8] Pyoluteorin biosynthesis begins with the activation of L-proline to prolyl-AMP by the adenylation domain PltF. With prolyl-AMP still in the active site, the active form of the peptidyl carrier protein PltL binds to PltF. Then PltF catalyzes the aminoacylation of PltL by attaching L-proline to the thiol of the 4’phosphopantetheine arm of PltL.[9] Next, the dehydrogenase PltE desaturates the prolyl moiety on PltL to create pyrrolyl-PltL. The halogenation domain PltA then dichlorinates the pyrrole moiety first at position 5 and then at position 4 in a FADH2 dependent manner.[10] The dichloropyrroyl residue is then transferred to the type I PKS PltB and PltC, however, the mechanism of transfer is unknown. The addition of 3 malonyl-CoA monomers, cyclization, and release by the thioesterase PltG gives pyoluteorin.
References
- ↑ "Genomics of secondary metabolite production by Pseudomonas spp". Natural Product Reports 26 (11): 1408–46. November 2009. doi:10.1039/b817075b. PMID 19844639.
- ↑ "Structure of a new antibiotic, pyoluteorin". Journal of the American Chemical Society 80 (17): 4749–4750. 1958. doi:10.1021/ja01550a093.
- ↑ "Influence of Enhanced Antibiotic Production in Pseudomonas fluorescens Strain CHA0 on its Disease Suppressive Capacity". Phytopathology 82 (2): 190–195. September 10, 1991. doi:10.1094/Phyto-82-190.
- ↑ "Suppression of Pythium ultimum-induced damping-off of cotton seedlings by pseudomonas fluorescens and its antibiotic, pyoluteorin". Phytopathology 70 (8): 712–715. January 16, 1980. doi:10.1094/Phyto-70-712.
- ↑ "Characterization of pyoluteorin derivatives as Mcl-1 antagonists". Cancer Research 74 (19): 1805. October 2014. doi:10.1158/1538-7445.AM2014-1805.
- ↑ "Biosynthesis of Pyoluteorin: A Mixed Polyketide-Tricarboxylic Acid Cycle Origin Demonstrated by [l,2-13C2]Acetate Incorporation". Zeitschrift für Naturforschung C 41 (5–6): 532–536. January 15, 1986. doi:10.1515/znc-1986-5-607.
- ↑ "Identification and sequence analysis of the genes encoding a polyketide synthase required for pyoluteorin biosynthesis in Pseudomonas fluorescens Pf-5". Gene 204 (1–2): 17–24. December 1997. doi:10.1016/S0378-1119(97)00501-5. PMID 9434161.
- ↑ "Characterization of the pyoluteorin biosynthetic gene cluster of Pseudomonas fluorescens Pf-5". Journal of Bacteriology 181 (7): 2166–74. April 1999. doi:10.1128/JB.181.7.2166-2174.1999. PMID 10094695.
- ↑ "Conversion of L-proline to pyrrolyl-2-carboxyl-S-PCP during undecylprodigiosin and pyoluteorin biosynthesis". Chemistry & Biology 9 (2): 171–84. February 2002. doi:10.1016/S1074-5521(02)00100-X. PMID 11880032.
- ↑ "Dichlorination of a pyrrolyl-S-carrier protein by FADH2-dependent halogenase PltA during pyoluteorin biosynthesis". Proceedings of the National Academy of Sciences of the United States of America 102 (39): 13843–8. September 2005. doi:10.1073/pnas.0506964102. PMID 16162666. Bibcode: 2005PNAS..10213843D.
 |
