Chemistry:Proline
| |||
| |||
| Names | |||
|---|---|---|---|
| IUPAC name
Proline
| |||
| Systematic IUPAC name
Pyrrolidine-2-carboxylic acid[1] | |||
| Identifiers | |||
3D model (JSmol)
|
|||
| 80812 | |||
| ChEBI |
| ||
| ChEMBL |
| ||
| ChemSpider | |||
| DrugBank |
| ||
| EC Number |
| ||
| 26927 | |||
| KEGG |
| ||
| MeSH | Proline | ||
PubChem CID
|
|||
| RTECS number |
| ||
| UNII |
| ||
| |||
| |||
| Properties | |||
| C5H9NO2 | |||
| Molar mass | 115.132 g·mol−1 | ||
| Appearance | Transparent crystals | ||
| Melting point | 205 to 228 °C (401 to 442 °F; 478 to 501 K) (decomposes) | ||
| Solubility | 1.5g/100g ethanol 19 degC[2] | ||
| log P | -0.06 | ||
| Acidity (pKa) | 1.99 (carboxyl), 10.96 (amino)[3] | ||
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |||
| Infobox references | |||
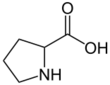
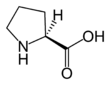
Proline (symbol Pro or P)[4] is an organic acid classed as a proteinogenic amino acid (used in the biosynthesis of proteins), although it does not contain the amino group -NH2 but is rather a secondary amine. The secondary amine nitrogen is in the protonated form (NH2+) under biological conditions, while the carboxyl group is in the deprotonated −COO− form. The "side chain" from the α carbon connects to the nitrogen forming a pyrrolidine loop, classifying it as a aliphatic amino acid. It is non-essential in humans, meaning the body can synthesize it from the non-essential amino acid L-glutamate. It is encoded by all the codons starting with CC (CCU, CCC, CCA, and CCG).
Proline is the only proteinogenic secondary amino acid which is a secondary amine, as the nitrogen atom is attached both to the α-carbon and to a chain of three carbons that together form a five-membered ring.
History and etymology
Proline was first isolated in 1900 by Richard Willstätter who obtained the amino acid while studying N-methylproline, and synthesized proline by the reaction of sodium salt of diethyl malonate with 1,3-dibromopropane. The next year, Emil Fischer isolated proline from casein and the decomposition products of γ-phthalimido-propylmalonic ester,[5] and published the synthesis of proline from phthalimide propylmalonic ester.[6]
The name proline comes from pyrrolidine, one of its constituents.[7]
Biosynthesis
Proline is biosynthetically derived from the amino acid L-glutamate. Glutamate-5-semialdehyde is first formed by glutamate 5-kinase (ATP-dependent) and glutamate-5-semialdehyde dehydrogenase (which requires NADH or NADPH). This can then either spontaneously cyclize to form 1-pyrroline-5-carboxylic acid, which is reduced to proline by pyrroline-5-carboxylate reductase (using NADH or NADPH), or turned into ornithine by ornithine aminotransferase, followed by cyclisation by ornithine cyclodeaminase to form proline.[8]
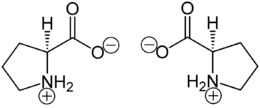
Biological activity
L-Proline has been found to act as a weak agonist of the glycine receptor and of both NMDA and non-NMDA (AMPA/kainate) ionotropic glutamate receptors.[9][10][11] It has been proposed to be a potential endogenous excitotoxin.[9][10][11] In plants, proline accumulation is a common physiological response to various stresses but is also part of the developmental program in generative tissues (e.g. pollen).[12][13][14][15]
A diet rich in proline was linked to an increased risk of depression in humans in a study from 2022 that was tested on a limited pre-clinical trial on humans and primarily in other organisms. Results were significant in the other organisms.[16]
Properties in protein structure
The distinctive cyclic structure of proline's side chain gives proline an exceptional conformational rigidity compared to other amino acids. It also affects the rate of peptide bond formation between proline and other amino acids. When proline is bound as an amide in a peptide bond, its nitrogen is not bound to any hydrogen, meaning it cannot act as a hydrogen bond donor, but can be a hydrogen bond acceptor.
Peptide bond formation with incoming Pro-tRNAPro in the ribosome is considerably slower than with any other tRNAs, which is a general feature of N-alkylamino acids.[17] Peptide bond formation is also slow between an incoming tRNA and a chain ending in proline; with the creation of proline-proline bonds slowest of all.[18]
The exceptional conformational rigidity of proline affects the secondary structure of proteins near a proline residue and may account for proline's higher prevalence in the proteins of thermophilic organisms. Protein secondary structure can be described in terms of the dihedral angles φ, ψ and ω of the protein backbone. The cyclic structure of proline's side chain locks the angle φ at approximately −65°.[19]
Proline acts as a structural disruptor in the middle of regular secondary structure elements such as alpha helices and beta sheets; however, proline is commonly found as the first residue of an alpha helix and also in the edge strands of beta sheets. Proline is also commonly found in turns (another kind of secondary structure), and aids in the formation of beta turns. This may account for the curious fact that proline is usually solvent-exposed, despite having a completely aliphatic side chain.
Multiple prolines and/or hydroxyprolines in a row can create a polyproline helix, the predominant secondary structure in collagen. The hydroxylation of proline by prolyl hydroxylase (or other additions of electron-withdrawing substituents such as fluorine) increases the conformational stability of collagen significantly.[20] Hence, the hydroxylation of proline is a critical biochemical process for maintaining the connective tissue of higher organisms. Severe diseases such as scurvy can result from defects in this hydroxylation, e.g., mutations in the enzyme prolyl hydroxylase or lack of the necessary ascorbate (vitamin C) cofactor.
Cis–trans isomerization
Peptide bonds to proline, and to other N-substituted amino acids (such as sarcosine), are able to populate both the cis and trans isomers. Most peptide bonds overwhelmingly adopt the trans isomer (typically 99.9% under unstrained conditions), chiefly because the amide hydrogen (trans isomer) offers less steric repulsion to the preceding Cα atom than does the following Cα atom (cis isomer). By contrast, the cis and trans isomers of the X-Pro peptide bond (where X represents any amino acid) both experience steric clashes with the neighboring substitution and have a much lower energy difference. Hence, the fraction of X-Pro peptide bonds in the cis isomer under unstrained conditions is significantly elevated, with cis fractions typically in the range of 3-10%.[21] However, these values depend on the preceding amino acid, with Gly[22] and aromatic[23] residues yielding increased fractions of the cis isomer. Cis fractions up to 40% have been identified for aromatic–proline peptide bonds.[24]
From a kinetic standpoint, cis–trans proline isomerization is a very slow process that can impede the progress of protein folding by trapping one or more proline residues crucial for folding in the non-native isomer, especially when the native protein requires the cis isomer. This is because proline residues are exclusively synthesized in the ribosome as the trans isomer form. All organisms possess prolyl isomerase enzymes to catalyze this isomerization, and some bacteria have specialized prolyl isomerases associated with the ribosome. However, not all prolines are essential for folding, and protein folding may proceed at a normal rate despite having non-native conformers of many X–Pro peptide bonds.
Uses
Proline and its derivatives are often used as asymmetric catalysts in proline organocatalysis reactions. The CBS reduction and proline catalysed aldol condensation are prominent examples.
In brewing, proteins rich in proline combine with polyphenols to produce haze (turbidity).[25]
L-Proline is an osmoprotectant and therefore is used in many pharmaceutical and biotechnological applications.
The growth medium used in plant tissue culture may be supplemented with proline. This can increase growth, perhaps because it helps the plant tolerate the stresses of tissue culture.[26][better source needed] For proline's role in the stress response of plants, see § Biological activity.
Specialties
Proline is one of the two amino acids that do not follow along with the typical Ramachandran plot, along with glycine. Due to the ring formation connected to the beta carbon, the ψ and φ angles about the peptide bond have fewer allowable degrees of rotation. As a result, it is often found in "turns" of proteins as its free entropy (ΔS) is not as comparatively large to other amino acids and thus in a folded form vs. unfolded form, the change in entropy is smaller. Furthermore, proline is rarely found in α and β structures as it would reduce the stability of such structures, because its side chain α-nitrogen can only form one nitrogen bond.
Additionally, proline is the only amino acid that does not form a red-purple colour when developed by spraying with ninhydrin for uses in chromatography. Proline, instead, produces an orange-yellow colour.
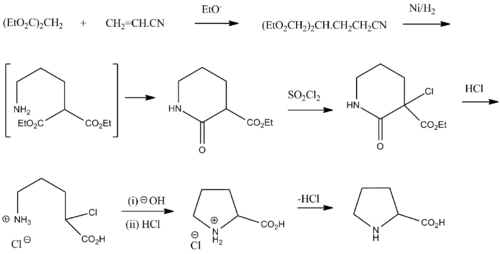
Synthesis
Racemic proline can be synthesized from diethyl malonate and acrylonitrile:[27]
See also
- Hyperprolinemia
- Inborn error of metabolism
- Prolidase deficiency
- Prolinol
References
- ↑ "Proline". PubChem. U.S. National Library of Medicine. https://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=614&loc=ec_rcs.
- ↑ Food Chemistry. 2009-01-15. p. 15. ISBN 978-3-540-69933-0. https://books.google.com/books?id=xteiARU46SQC&q=proline&pg=PA15.
- ↑ Principles of Biochemistry. New York: W.H. Freeman and Company.
- ↑ "Nomenclature and Symbolism for Amino Acids and Peptides". IUPAC-IUB Joint Commission on Biochemical Nomenclature. 1983. http://www.chem.qmul.ac.uk/iupac/AminoAcid/AA1n2.html.
- ↑ The chemical composition of the proteins, Monographs on biochemistry, Part I. Analysis (2nd ed.), London: Longmans, Green and Co., 1912, p. 130, https://books.google.com/books?id=7JM8AAAAIAAJ&pg=PA130, retrieved September 20, 2010
- ↑ "Proline". Amino Acids Guide. http://www.aminoacidsguide.com/Pro.html.
- ↑ "Proline". American Heritage Dictionary of the English Language, 4th edition. http://www.yourdictionary.com/proline.
- ↑ Lehninger, Albert L.; Nelson, David L.; Cox, Michael M. (2000). Principles of Biochemistry (3rd ed.). New York: W. H. Freeman. ISBN 1-57259-153-6..
- ↑ 9.0 9.1 Ion Channel Factsbook: Extracellular Ligand-Gated Channels. Academic Press. 16 November 1995. pp. 126. ISBN 978-0-08-053519-7. https://books.google.com/books?id=XqzdupxdZUgC&pg=PA126.
- ↑ 10.0 10.1 "L-proline activates glutamate and glycine receptors in cultured rat dorsal horn neurons". Molecular Pharmacology 41 (4): 793–801. April 1992. PMID 1349155. http://molpharm.aspetjournals.org/cgi/pmidlookup?view=long&pmid=1349155.
- ↑ 11.0 11.1 Neuroanatomical Basis of Clinical Neurology (Second ed.). CRC Press. 7 August 2014. pp. 309. ISBN 978-1-4398-4833-3. https://books.google.com/books?id=UTv6AwAAQBAJ&pg=PA309.
- ↑ "Proline accumulation in plants: a review". Amino Acids 35 (4): 753–759. November 2008. doi:10.1007/s00726-008-0061-6. PMID 18379856.
- ↑ "Natural diversity uncovers P5CS1 regulation and its role in drought stress tolerance and yield sustainability in barley". Plant, Cell & Environment 45 (12): 3523–3536. September 2022. doi:10.1111/pce.14445. PMID 36130879.
- ↑ "Abscisic acid-responsive element binding transcription factors contribute to proline synthesis and stress adaptation in Arabidopsis". Journal of Plant Physiology 261: 153414. June 2021. doi:10.1016/j.jplph.2021.153414. PMID 33895677.
- ↑ "An Ancestral Allele of Pyrroline-5-carboxylate synthase1 Promotes Proline Accumulation and Drought Adaptation in Cultivated Barley". Plant Physiology 178 (2): 771–782. October 2018. doi:10.1104/pp.18.00169. PMID 30131422.
- ↑ "Microbiota alterations in proline metabolism impact depression". Cell Metabolism 34 (5): 681–701.e10. May 2022. doi:10.1016/j.cmet.2022.04.001. PMID 35508109.
- ↑ "Slow peptide bond formation by proline and other N-alkylamino acids in translation". Proceedings of the National Academy of Sciences of the United States of America 106 (1): 50–54. January 2009. doi:10.1073/pnas.0809211106. PMID 19104062. Bibcode: 2009PNAS..106...50P..
- ↑ "Biochemistry. Getting past polyproline pauses". Science 339 (6115): 38–39. January 2013. doi:10.1126/science.1233338. PMID 23288527. Bibcode: 2013Sci...339...38B.
- ↑ "Stereochemical quality of protein structure coordinates". Proteins 12 (4): 345–364. April 1992. doi:10.1002/prot.340120407. PMID 1579569.
- ↑ "Fish bone chemistry and ultrastructure: implications for taphonomy and stable isotope analysis". Journal of Archaeological Science 38 (12): 3358–3372. 2011. doi:10.1016/j.jas.2011.07.022. https://uwo.academia.edu/PaulSzpak/Papers/827788/Fish_Bone_Chemistry_and_Ultrastructure_Implications_for_Taphonomy_and_Stable_Isotope_Analysis.
- ↑ "Propensity for cis-Proline Formation in Unfolded Proteins". ChemBioChem 19 (1): 37–42. January 2018. doi:10.1002/cbic.201700548. PMID 29064600.
- ↑ "Detection of cis and trans X–Pro peptide bonds in proteins by 13C NMR: application to collagen". Proceedings of the National Academy of Sciences of the United States of America 81 (15): 4800–4803. August 1984. doi:10.1073/pnas.81.15.4800. PMID 6589627. Bibcode: 1984PNAS...81.4800S.
- ↑ "Electronic control of amide cis–trans isomerism via the aromatic-prolyl interaction". Journal of the American Chemical Society 128 (7): 2216–2217. February 2006. doi:10.1021/ja057901y. PMID 16478167.
- ↑ "A Slow Conformational Switch in the BMAL1 Transactivation Domain Modulates Circadian Rhythms". Molecular Cell 66 (4): 447–457.e7. May 2017. doi:10.1016/j.molcel.2017.04.011. PMID 28506462.
- ↑ "Haze and Foam". http://www.nysaes.cornell.edu/fst/faculty/siebert/haze.html. Accessed July 12, 2010.
- ↑ "Effects of Some Organic Nitrogen Sources and Antibiotics on Callus Growth of Indica Rice Cultivars". Journal of Plant Nutrition 38 (8): 1231–1240. 2015. doi:10.1080/01904167.2014.983118.
- ↑ Vogel, Practical Organic Chemistry 5th edition
Further reading
- "Proline isomerization and its catalysis in protein folding". Mechanisms of Protein Folding (2nd ed.). Oxford University Press. 2000. pp. 212–249. ISBN 978-0-19-963788-1..
External links
 |