Latent Dirichlet allocation
In natural language processing, latent Dirichlet allocation (LDA) is a Bayesian network (and, therefore, a generative statistical model) for modeling automatically extracted topics in textual corpora. The LDA is an example of a Bayesian topic model. In this, observations (e.g., words) are collected into documents, and each word's presence is attributable to one of the document's topics. Each document will contain a small number of topics.
History
In the context of population genetics, LDA was proposed by J. K. Pritchard, M. Stephens and P. Donnelly in 2000.[1][2]
LDA was applied in machine learning by David Blei, Andrew Ng and Michael I. Jordan in 2003.[3]
Overview
Evolutionary biology and bio-medicine
In evolutionary biology and bio-medicine, the model is used to detect the presence of structured genetic variation in a group of individuals. The model assumes that alleles carried by individuals under study have origin in various extant or past populations. The model and various inference algorithms allow scientists to estimate the allele frequencies in those source populations and the origin of alleles carried by individuals under study. The source populations can be interpreted ex-post in terms of various evolutionary scenarios. In association studies, detecting the presence of genetic structure is considered a necessary preliminary step to avoid confounding.
Clinical psychology, mental health, and social science
In clinical psychology research, LDA has been used to identify common themes of self-images experienced by young people in social situations.[4] Other social scientists have used LDA to examine large sets of topical data from discussions on social media (e.g., tweets about prescription drugs).[5]
Musicology
In the context of computational musicology, LDA has been used to discover tonal structures in different corpora.[6]
Machine learning
One application of LDA in machine learning - specifically, topic discovery, a subproblem in natural language processing – is to discover topics in a collection of documents, and then automatically classify any individual document within the collection in terms of how "relevant" it is to each of the discovered topics. A topic is considered to be a set of terms (i.e., individual words or phrases) that, taken together, suggest a shared theme.
For example, in a document collection related to pet animals, the terms dog, spaniel, beagle, golden retriever, puppy, bark, and woof would suggest a DOG_related theme, while the terms cat, siamese, Maine coon, tabby, manx, meow, purr, and kitten would suggest a CAT_related theme. There may be many more topics in the collection – e.g., related to diet, grooming, healthcare, behavior, etc. that we do not discuss for simplicity's sake. (Very common, so called stop words in a language – e.g., "the", "an", "that", "are", "is", etc., – would not discriminate between topics and are usually filtered out by pre-processing before LDA is performed. Pre-processing also converts terms to their "root" lexical forms – e.g., "barks", "barking", and "barked" would be converted to "bark".)
If the document collection is sufficiently large, LDA will discover such sets of terms (i.e., topics) based upon the co-occurrence of individual terms, though the task of assigning a meaningful label to an individual topic (i.e., that all the terms are DOG_related) is up to the user, and often requires specialized knowledge (e.g., for collection of technical documents). The LDA approach assumes that:
- The semantic content of a document is composed by combining one or more terms from one or more topics.
- Certain terms are ambiguous, belonging to more than one topic, with different probability. (For example, the term training can apply to both dogs and cats, but are more likely to refer to dogs, which are used as work animals or participate in obedience or skill competitions.) However, in a document, the accompanying presence of specific neighboring terms (which belong to only one topic) will disambiguate their usage.
- Most documents will contain only a relatively small number of topics. In the collection, e.g., individual topics will occur with differing frequencies. That is, they have a probability distribution, so that a given document is more likely to contain some topics than others.
- Within a topic, certain terms will be used much more frequently than others. In other words, the terms within a topic will also have their own probability distribution.
When LDA machine learning is employed, both sets of probabilities are computed during the training phase, using Bayesian methods and an Expectation Maximization algorithm.
LDA is a generalization of older approach of probabilistic latent semantic analysis (pLSA), The pLSA model is equivalent to LDA under a uniform Dirichlet prior distribution.[7] pLSA relies on only the first two assumptions above and does not care about the remainder. While both methods are similar in principle and require the user to specify the number of topics to be discovered before the start of training (as with K-means clustering) LDA has the following advantages over pLSA:
- LDA yields better disambiguation of words and a more precise assignment of documents to topics.
- Computing probabilities allows a "generative" process by which a collection of new “synthetic documents” can be generated that would closely reflect the statistical characteristics of the original collection.
- Unlike LDA, pLSA is vulnerable to overfitting especially when the size of corpus increases.
- The LDA algorithm is more readily amenable to scaling up for large data sets using the MapReduce approach on a computing cluster.
Model

With plate notation, which is often used to represent probabilistic graphical models (PGMs), the dependencies among the many variables can be captured concisely. The boxes are "plates" representing replicates, which are repeated entities. The outer plate represents documents, while the inner plate represents the repeated word positions in a given document; each position is associated with a choice of topic and word. The variable names are defined as follows:
- M denotes the number of documents
- N is number of words in a given document (document i has words)
- α is the parameter of the Dirichlet prior on the per-document topic distributions
- β is the parameter of the Dirichlet prior on the per-topic word distribution
- is the topic distribution for document i
- is the word distribution for topic k
- is the topic for the j-th word in document i
- is the specific word.
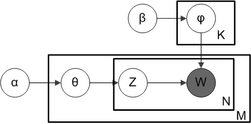
The fact that W is grayed out means that words are the only observable variables, and the other variables are latent variables. As proposed in the original paper,[3] a sparse Dirichlet prior can be used to model the topic-word distribution, following the intuition that the probability distribution over words in a topic is skewed, so that only a small set of words have high probability. The resulting model is the most widely applied variant of LDA today. The plate notation for this model is shown on the right, where denotes the number of topics and are -dimensional vectors storing the parameters of the Dirichlet-distributed topic-word distributions ( is the number of words in the vocabulary).
It is helpful to think of the entities represented by and as matrices created by decomposing the original document-word matrix that represents the corpus of documents being modeled. In this view, consists of rows defined by documents and columns defined by topics, while consists of rows defined by topics and columns defined by words. Thus, refers to a set of rows, or vectors, each of which is a distribution over words, and refers to a set of rows, each of which is a distribution over topics.
Generative process
To actually infer the topics in a corpus, we imagine a generative process whereby the documents are created, so that we may infer, or reverse engineer, it. We imagine the generative process as follows. Documents are represented as random mixtures over latent topics, where each topic is characterized by a distribution over all the words. LDA assumes the following generative process for a corpus consisting of documents each of length :
1. Choose , where and is a Dirichlet distribution with a symmetric parameter which typically is sparse ()
2. Choose , where and typically is sparse
3. For each of the word positions , where , and
- (a) Choose a topic
- (b) Choose a word
(Note that multinomial distribution here refers to the multinomial with only one trial, which is also known as the categorical distribution.)
The lengths are treated as independent of all the other data generating variables ( and ). The subscript is often dropped, as in the plate diagrams shown here.
Definition
A formal description of LDA is as follows:
| Variable | Type | Meaning |
|---|---|---|
| integer | number of topics (e.g. 50) | |
| integer | number of words in the vocabulary (e.g. 50,000 or 1,000,000) | |
| integer | number of documents | |
| integer | number of words in document d | |
| integer | total number of words in all documents; sum of all values, i.e. | |
| positive real | prior weight of topic k in a document; usually the same for all topics; normally a number less than 1, e.g. 0.1, to prefer sparse topic distributions, i.e. few topics per document | |
| K-dimensional vector of positive reals | collection of all values, viewed as a single vector | |
| positive real | prior weight of word w in a topic; usually the same for all words; normally a number much less than 1, e.g. 0.001, to strongly prefer sparse word distributions, i.e. few words per topic | |
| V-dimensional vector of positive reals | collection of all values, viewed as a single vector | |
| probability (real number between 0 and 1) | probability of word w occurring in topic k | |
| V-dimensional vector of probabilities, which must sum to 1 | distribution of words in topic k | |
| probability (real number between 0 and 1) | probability of topic k occurring in document d | |
| K-dimensional vector of probabilities, which must sum to 1 | distribution of topics in document d | |
| integer between 1 and K | identity of topic of word w in document d | |
| N-dimensional vector of integers between 1 and K | identity of topic of all words in all documents | |
| integer between 1 and V | identity of word w in document d | |
| N-dimensional vector of integers between 1 and V | identity of all words in all documents |
We can then mathematically describe the random variables as follows:
Inference
Learning the various distributions (the set of topics, their associated word probabilities, the topic of each word, and the particular topic mixture of each document) is a problem of statistical inference.
Monte Carlo simulation
The original paper by Pritchard et al.[1] used approximation of the posterior distribution by Monte Carlo simulation. Alternative proposal of inference techniques include Gibbs sampling.[8]
Variational Bayes
The original ML paper used a variational Bayes approximation of the posterior distribution.[3]
Likelihood maximization
A direct optimization of the likelihood with a block relaxation algorithm proves to be a fast alternative to MCMC.[9]
Unknown number of populations/topics
In practice, the optimal number of populations or topics is not known beforehand. It can be estimated by approximation of the posterior distribution with reversible-jump Markov chain Monte Carlo.[10]
Alternative approaches
Alternative approaches include expectation propagation.[11]
Recent research has been focused on speeding up the inference of latent Dirichlet allocation to support the capture of a massive number of topics in a large number of documents. The update equation of the collapsed Gibbs sampler mentioned in the earlier section has a natural sparsity within it that can be taken advantage of. Intuitively, since each document only contains a subset of topics , and a word also only appears in a subset of topics , the above update equation could be rewritten to take advantage of this sparsity.[12]
In this equation, we have three terms, out of which two are sparse, and the other is small. We call these terms and respectively. Now, if we normalize each term by summing over all the topics, we get:
Here, we can see that is a summation of the topics that appear in document , and is also a sparse summation of the topics that a word is assigned to across the whole corpus. on the other hand, is dense but because of the small values of & , the value is very small compared to the two other terms.
Now, while sampling a topic, if we sample a random variable uniformly from , we can check which bucket our sample lands in. Since is small, we are very unlikely to fall into this bucket; however, if we do fall into this bucket, sampling a topic takes time (same as the original Collapsed Gibbs Sampler). However, if we fall into the other two buckets, we only need to check a subset of topics if we keep a record of the sparse topics. A topic can be sampled from the bucket in time, and a topic can be sampled from the bucket in time where and denotes the number of topics assigned to the current document and current word type respectively.
Notice that after sampling each topic, updating these buckets is all basic arithmetic operations.
Aspects of computational details
Following is the derivation of the equations for collapsed Gibbs sampling, which means s and s will be integrated out. For simplicity, in this derivation the documents are all assumed to have the same length . The derivation is equally valid if the document lengths vary.
According to the model, the total probability of the model is:
where the bold-font variables denote the vector version of the variables. First, and need to be integrated out.
All the s are independent to each other and the same to all the s. So we can treat each and each separately. We now focus only on the part.
We can further focus on only one as the following:
Actually, it is the hidden part of the model for the document. Now we replace the probabilities in the above equation by the true distribution expression to write out the explicit equation.
Let be the number of word tokens in the document with the same word symbol (the word in the vocabulary) assigned to the topic. So, is three dimensional. If any of the three dimensions is not limited to a specific value, we use a parenthesized point to denote. For example, denotes the number of word tokens in the document assigned to the topic. Thus, the right most part of the above equation can be rewritten as:
So the integration formula can be changed to:
The equation inside the integration has the same form as the Dirichlet distribution. According to the Dirichlet distribution,
Thus,
Now we turn our attention to the part. Actually, the derivation of the part is very similar to the part. Here we only list the steps of the derivation:
For clarity, here we write down the final equation with both and integrated out:
The goal of Gibbs Sampling here is to approximate the distribution of . Since is invariable for any of Z, Gibbs Sampling equations can be derived from directly. The key point is to derive the following conditional probability:
where denotes the hidden variable of the word token in the document. And further we assume that the word symbol of it is the word in the vocabulary. denotes all the s but . Note that Gibbs Sampling needs only to sample a value for , according to the above probability, we do not need the exact value of
but the ratios among the probabilities that can take value. So, the above equation can be simplified as:
Finally, let be the same meaning as but with the excluded. The above equation can be further simplified leveraging the property of gamma function. We first split the summation and then merge it back to obtain a -independent summation, which could be dropped:
Note that the same formula is derived in the article on the Dirichlet-multinomial distribution, as part of a more general discussion of integrating Dirichlet distribution priors out of a Bayesian network.
Related problems
Related models
Topic modeling is a classic solution to the problem of information retrieval using linked data and semantic web technology.[13] Related models and techniques are, among others, latent semantic indexing, independent component analysis, probabilistic latent semantic indexing, non-negative matrix factorization, and Gamma-Poisson distribution.
The LDA model is highly modular and can therefore be easily extended. The main field of interest is modeling relations between topics. This is achieved by using another distribution on the simplex instead of the Dirichlet. The Correlated Topic Model[14] follows this approach, inducing a correlation structure between topics by using the logistic normal distribution instead of the Dirichlet. Another extension is the hierarchical LDA (hLDA),[15] where topics are joined together in a hierarchy by using the nested Chinese restaurant process, whose structure is learnt from data. LDA can also be extended to a corpus in which a document includes two types of information (e.g., words and names), as in the LDA-dual model.[16] Nonparametric extensions of LDA include the hierarchical Dirichlet process mixture model, which allows the number of topics to be unbounded and learnt from data.
As noted earlier, pLSA is similar to LDA. The LDA model is essentially the Bayesian version of pLSA model. The Bayesian formulation tends to perform better on small datasets because Bayesian methods can avoid overfitting the data. For very large datasets, the results of the two models tend to converge. One difference is that pLSA uses a variable to represent a document in the training set. So in pLSA, when presented with a document the model has not seen before, we fix —the probability of words under topics—to be that learned from the training set and use the same EM algorithm to infer —the topic distribution under . Blei argues that this step is cheating because you are essentially refitting the model to the new data.
Spatial models
In evolutionary biology, it is often natural to assume that the geographic locations of the individuals observed bring some information about their ancestry. This is the rational of various models for geo-referenced genetic data.[10][17]
Variations on LDA have been used to automatically put natural images into categories, such as "bedroom" or "forest", by treating an image as a document, and small patches of the image as words;[18] one of the variations is called spatial latent Dirichlet allocation.[19]
See also
References
- ↑ 1.0 1.1 Pritchard, J. K.; Stephens, M.; Donnelly, P. (June 2000). "Inference of population structure using multilocus genotype data.". Genetics 155 (2): pp. 945–959. doi:10.1093/genetics/155.2.945. ISSN 0016-6731. PMID 10835412. PMC 1461096. http://genetics.org/content/155/2/945.
- ↑ Falush, D.; Stephens, M.; Pritchard, J. K. (2003). "Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies.". Genetics 164 (4): pp. 1567–1587. doi:10.1093/genetics/164.4.1567. PMID 12930761.
- ↑ 3.0 3.1 3.2 Blei, David M.; Ng, Andrew Y.; Jordan, Michael I (January 2003). Lafferty, John. ed. "Latent Dirichlet Allocation". Journal of Machine Learning Research 3 (4–5): pp. 993–1022. doi:10.1162/jmlr.2003.3.4-5.993. http://jmlr.csail.mit.edu/papers/v3/blei03a.html. Retrieved 2006-12-19.
- ↑ Chiu, Kin; Clark, David; Leigh, Eleanor (July 2022). "Characterising Negative Mental Imagery in Adolescent Social Anxiety". Cognitive Therapy and Research 46 (5): 956–966. doi:10.1007/s10608-022-10316-x. PMID 36156987.
- ↑ Parker, Maria A.; Valdez, Danny; Rao, Varun K.; Eddens, Katherine S.; Agley, Jon (2023). "Results and Methodological Implications of the Digital Epidemiology of Prescription Drug References Among Twitter Users: Latent Dirichlet Allocation (LDA) Analyses" (in EN). Journal of Medical Internet Research 25 (1): e48405. doi:10.2196/48405. PMID 37505795.
- ↑ Lieck, Robert; Moss, Fabian C.; Rohrmeier, Martin (October 2020). "The Tonal Diffusion Model". Transactions of the International Society for Music Information Retrieval 3 (1): pp. 153–164. doi:10.5334/tismir.46.
- ↑ Girolami, Mark; Kaban, A. (2003). "On an Equivalence between PLSI and LDA". Proceedings of SIGIR 2003. New York: Association for Computing Machinery. ISBN 1-58113-646-3. https://archive.org/details/sigir2003proceed0000inte.
- ↑ Griffiths, Thomas L.; Steyvers, Mark (April 6, 2004). "Finding scientific topics". Proceedings of the National Academy of Sciences 101 (Suppl. 1): 5228–5235. doi:10.1073/pnas.0307752101. PMID 14872004. Bibcode: 2004PNAS..101.5228G.
- ↑ Alexander, David H.; Novembre, John; Lange, Kenneth (2009). "Fast model-based estimation of ancestry in unrelated individuals". Genome Research 19 (9): 1655–1664. doi:10.1101/gr.094052.109. PMID 19648217.
- ↑ 10.0 10.1 Guillot, G.; Estoup, A.; Mortier, F.; Cosson, J. (2005). "A spatial statistical model for landscape genetics". Genetics 170 (3): pp. 1261–1280. doi:10.1534/genetics.104.033803. PMID 15520263.
- ↑ Minka, Thomas; Lafferty, John (2002). "Expectation-propagation for the generative aspect model". Proceedings of the 18th Conference on Uncertainty in Artificial Intelligence. San Francisco, CA: Morgan Kaufmann. ISBN 1-55860-897-4. https://research.microsoft.com/~minka/papers/aspect/minka-aspect.pdf.
- ↑ Yao, Limin; Mimno, David; McCallum, Andrew (2009). "Efficient methods for topic model inference on streaming document collections". 15th ACM SIGKDD international conference on Knowledge discovery and data mining.
- ↑ Lamba, Manika; Madhusudhan, Margam (2019). "Mapping of topics in DESIDOC Journal of Library and Information Technology, India: a study". Scientometrics 120 (2): 477–505. doi:10.1007/s11192-019-03137-5.
- ↑ Blei, David M.; Lafferty, John D. (2005). "Correlated topic models". Advances in Neural Information Processing Systems 18. https://papers.nips.cc/paper/2005/file/9e82757e9a1c12cb710ad680db11f6f1-Paper.pdf.
- ↑ Blei, David M.; Jordan, Michael I.; Griffiths, Thomas L.; Tenenbaum, Joshua B (2004). "Hierarchical Topic Models and the Nested Chinese Restaurant Process". Advances in Neural Information Processing Systems 16: Proceedings of the 2003 Conference. MIT Press. ISBN 0-262-20152-6. https://proceedings.neurips.cc/paper/2003/file/7b41bfa5085806dfa24b8c9de0ce567f-Paper.pdf.
- ↑ Shu, Liangcai; Long, Bo; Meng, Weiyi (2009). "A Latent Topic Model for Complete Entity Resolution". 25th IEEE International Conference on Data Engineering (ICDE 2009). http://www.cs.binghamton.edu/~meng/pub.d/icde09-LatentTopic.pdf.
- ↑ Guillot, G.; Leblois, R.; Coulon, A.; Frantz, A. (2009). "Statistical methods in spatial genetics". Molecular Ecology 18 (23): pp. 4734–4756. doi:10.1111/j.1365-294X.2009.04410.x. PMID 19878454.
- ↑ Li, Fei-Fei; Perona, Pietro. "A Bayesian Hierarchical Model for Learning Natural Scene Categories". Proceedings of the 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR'05) 2: 524–531.
- ↑ Wang, Xiaogang; Grimson, Eric (2007). "Spatial Latent Dirichlet Allocation". Proceedings of Neural Information Processing Systems Conference (NIPS). https://papers.nips.cc/paper/2007/file/ec8956637a99787bd197eacd77acce5e-Paper.pdf.
External links
- jLDADMM A Java package for topic modeling on normal or short texts. jLDADMM includes implementations of the LDA topic model and the one-topic-per-document Dirichlet Multinomial Mixture model. jLDADMM also provides an implementation for document clustering evaluation to compare topic models.
- STTM A Java package for short text topic modeling (https://github.com/qiang2100/STTM). STTM includes these following algorithms: Dirichlet Multinomial Mixture (DMM) in conference KDD2014, Biterm Topic Model (BTM) in journal TKDE2016, Word Network Topic Model (WNTM) in journal KAIS2018, Pseudo-Document-Based Topic Model (PTM) in conference KDD2016, Self-Aggregation-Based Topic Model (SATM) in conference IJCAI2015, (ETM) in conference PAKDD2017, Generalized P´olya Urn (GPU) based Dirichlet Multinomial Mixturemodel (GPU-DMM) in conference SIGIR2016, Generalized P´olya Urn (GPU) based Poisson-based Dirichlet Multinomial Mixturemodel (GPU-PDMM) in journal TIS2017 and Latent Feature Model with DMM (LF-DMM) in journal TACL2015. STTM also includes six short text corpus for evaluation. STTM presents three aspects about how to evaluate the performance of the algorithms (i.e., topic coherence, clustering, and classification).
- Lecture that covers some of the notation in this article: LDA and Topic Modelling Video Lecture by David Blei or same lecture on YouTube
- D. Mimno's LDA Bibliography An exhaustive list of LDA-related resources (incl. papers and some implementations)
- Gensim, a Python+NumPy implementation of online LDA for inputs larger than the available RAM.
- topicmodels and lda are two R packages for LDA analysis.
- MALLET Open source Java-based package from the University of Massachusetts-Amherst for topic modeling with LDA, also has an independently developed GUI, the Topic Modeling Tool
- LDA in Mahout implementation of LDA using MapReduce on the Hadoop platform
- Latent Dirichlet Allocation (LDA) Tutorial for the Infer.NET Machine Computing Framework Microsoft Research C# Machine Learning Framework
- LDA in Spark: Since version 1.3.0, Apache Spark also features an implementation of LDA
- LDA, exampleLDA MATLAB implementation
 |
