Medicine:Studierfenster
The Studierfenster Logo | |
| Developer(s) | University Hospital Essen, Graz University of Technology, Medical University of Graz |
|---|---|
| Initial release | 2018 |
| Written in | C, C++, Python, JavaScript, HTML |
| Operating system | Cross-platform (Windows, Mac OS X, Linux) |
| Available in | English |
| Type | Image processing, scientific visualization, medical imaging, volume rendering, Interactive visualization |
| License | GPL, CC BY-SA |
| Website | studierfenster |
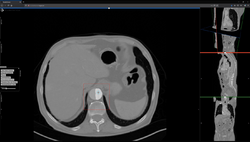
Studierfenster or StudierFenster (SF)[1][2][3] is a free, non-commercial open science client/server-based medical imaging processing online framework. It offers capabilities, like viewing medical data (computed tomography (CT), magnetic resonance imaging (MRI), etc.) in two- and three-dimensional space directly in the standard web browsers, like Google Chrome, Mozilla Firefox, Safari, and Microsoft Edge. Other functionalities are the calculation of medical metrics (dice score[4] and Hausdorff distance[5]), manual slice-by-slice outlining of structures in medical images (segmentation[6][7]), manual placing of (anatomical) landmarks in medical image data, viewing medical data in virtual reality, a facial reconstruction and registration of medical data for augmented reality,[8] one click showcases for COVID-19 and veterinary scans, and a Radiomics module.
Other features of Studierfenster are the automatic cranial implant design with a neural network,[9][10] the inpainting of aortic dissections[11] with a generative adversarial network,[12][13] an automatic aortic landmark detection with deep learning[14] in computed tomography angiography scans, and a GrowCut algorithm implementation for image segmentation.
Studierfenster is currently hosted on a server at the Graz University of Technology[15] in Austria, and expanded jointly with the Institute for Artificial Intelligence in Medicine (IKIM)[16] in Essen, Germany.
History
Studierfenster was initiated within two bachelor theses during the summer bachelor program of the Institute of Computer Graphics and Vision (ICG) at Graz University of Technology, Austria, in cooperation with the Medical University of Graz, Austria, in 2018/2019.[17][18]
The name Studierfenster (or StudierFenster) is German and can be translated to 'StudyWindow', whereby window refers here to a browser window. The word Studierfenster is an adaption from the word Studierstube ('study room'), which was an augmented reality project at the Vienna University of Technology in Austria.[19][20]
Architecture
Studierfenster is set up as a distributed application via a client–server model. The client side (front-end) consists of HTML and JavaScript with WebGL to enable 2D and 3D visualization, rendered on the client.
The server side (back-end) handles client requests via C, C++ and Python.[21] It interfaces to common open source libraries and software tools like the Insight Toolkit,[22] the Visualization Toolkit (VTK),[23] the X Toolkit (XTK)[24] and Slice:Drop.[25] The server communication is handled by AJAX requests[26] were needed.
Studierfenster employs a Flask server.
Features
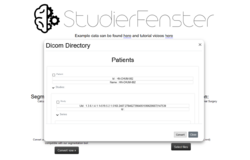
Dicom browser
This allows client-side parsing a local folder with DICOM (Digital Imaging and Communications in Medicine)[27][28] files. Afterwards, the whole folder can be converted to compressed .nrrd (nearly raw raster data) files and downloaded as a single .zip file.
Nrrd is a library and file format for the representation and processing of n-dimensional raster data. It is intended to support scientific visualization and (medical) image processing applications.[29] With the "Dicom Browser" of Studierfenster, it is possible to select specific Studies or Series, and only convert these.
File converter
The file converter converts a medical volume file (e.g. a non-compressed .nrrd file) to a compressed/binary .nrrd file. After the conversion, the compressed .nrrd file can be downloaded and used with the "Medical 3D Viewer" for 2D and 3D visualization, and further image processing.
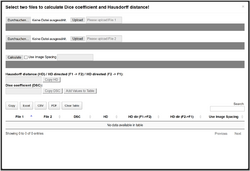
Metrics module
This can calculate the Dice similarity coefficient and Hausdorff distance between two segmentation masks (in .nrrd format) in a standard web browser.
The resulting table has seven columns: the file names for both files used in the calculation, the calculated Dice similarity coefficient, the calculated Hausdorff distance, the calculated directed HD for both directions, and the information if image spacing was used in the calculation. The table can be sorted, is searchable, and can be exported as a simple copy, an Excel spreadsheet, a comma-separated values file or as a portable document format.
The Metrics Module has been used to compare manual anatomical segmentations of brain tumors[30]
VR viewer
The VR Viewer (or Medical VR Viewer) enables viewing (medical) data in Virtual Reality (VR) with devices like the Google Cardboard or the HTC Vive (via the WebVR App).[31] For viewing the data in VR, it needs to be converted to the VTI (.vti) format, which can be done with open-source, multi-platform data analysis and visualization application ParaView[32]
Critics
Studierfenster is not a certified medical product; it can only be used for educational, research, and informational purposes.
References
- ↑ "Studierfenster". http://www.studierfenster.at.
- ↑ Egger, Jan; Wild, Daniel; Weber, Maximilian; Ramirez Bedoya, Christopher; Karner, Florian; Prutsch, Alexander; Schmied, Michael; Dionysio, Christina et al. (2022). "Studierfenster: an Open Science Cloud-Based Medical Imaging Analysis Platform" (in en). Journal of Digital Imaging (Cham: Springer International Publishing) 35 (2): 340–355. doi:10.1007/s10278-021-00574-8. PMID 35064372.
- ↑ Weber, Maximilian (2019-10-17). "A Client/Server based Online Environment for the Calculation of Medical Segmentation Scores" (in en-US). 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). 2019. pp. 3463–3467. doi:10.1109/EMBC.2019.8856481. ISBN 978-1-5386-1311-5.
- ↑ Dice, Lee R. (1945). "Measures of the Amount of Ecologic Association Between Species". Ecology 26 (3): 297–302. doi:10.2307/1932409. Bibcode: 1945Ecol...26..297D.
- ↑ Rockafellar, R. Tyrrell; Wets, Roger J-B (2005). Variational Analysis. Springer-Verlag. p. 117. ISBN 3-540-62772-3.
- ↑ Linda G. Shapiro and George C. Stockman (2001): “Computer Vision”, pp 279–325, New Jersey, Prentice-Hall, ISBN:0-13-030796-3
- ↑ Barghout, Lauren, and Lawrence W. Lee. "Perceptual information processing system." Paravue Inc. U.S. Patent Application 10/618,543, filed July 11, 2003.
- ↑ Gsaxner, Christina; Pepe, Antonio; Wallner, Jürgen; Schmalstieg, Dieter; Egger, Jan (2019). "Markerless Image-to-Face Registration for Untethered Augmented Reality in Head and Neck Surgery". in Shen, Dinggang; Liu, Tianming; Peters, Terry M. et al. (in en). Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. Lecture Notes in Computer Science. 11768. Cham: Springer International Publishing. pp. 236–244. doi:10.1007/978-3-030-32254-0_27. ISBN 978-3-030-32254-0.
- ↑ Li, Jianning (January 2020). Deep Learning for Cranial Defect Reconstruction. Master Thesis, Institute of Computer Graphics and Vision, Graz University of Technology, Austria, pp. 1-68, January 2020. (Master's Thesis).
- ↑ Li, Jianning; Pepe, Antonio; Gsaxner, Christina; Egger, Jan (2021). "An online platform for automatic skull defect restoration and cranial implant design". Medical Imaging 2021: Image-Guided Procedures, Robotic Interventions, and Modeling. 11598. 59. doi:10.1117/12.2580719. ISBN 9781510640252. Bibcode: 2021SPIE11598E..1QL.
- ↑ Pepe, Antonio; Li, Jianning; Rolf-Pissarczyk, Malte; Gsaxner, Christina; Chen, Xiaojun; Holzapfel, Gerhard A.; Egger, Jan (2020). "Detection, Segmentation, Simulation and Visualization of Aortic Dissections: A Review". Medical Image Analysis 65: 101773. doi:10.1016/j.media.2020.101773. PMID 32738647.
- ↑ Prutsch, Alexander. "Design and Development of a Web-based Tool for Inpainting ofDissected Aortae in Angiography Images". https://cescg.org/wp-content/uploads/2020/03/Prutsch-Design-and-Development-of-a-Web-based-Tool-for-Inpainting-of-Dissected-Aortae-in-Angiography-Images.pdf.
- ↑ Goodfellow, Ian; Pouget-Abadie, Jean; Mirza, Mehdi; Xu, Bing; Warde-Farley, David; Ozair, Sherjil; Courville, Aaron; Bengio, Yoshua (2014). "Generative Adversarial Networks". Proceedings of the International Conference on Neural Information Processing Systems (NIPS 2014). pp. 2672–2680. https://papers.nips.cc/paper/5423-generative-adversarial-nets.pdf.
- ↑ Schmidhuber, J. (2015). "Deep Learning in Neural Networks: An Overview". Neural Networks 61: 85–117. doi:10.1016/j.neunet.2014.09.003. PMID 25462637.
- ↑ "Graz University of Technology". https://www.tugraz.at/.
- ↑ "Artificial Intelligence in Medicine". https://www.ikim.uk-essen.de/.
- ↑ Weber, Maximilian (2018-12-13). A Client/Server based Online Environment for the calculation of Segmentation Scores (Bachelor Thesis). Austria: Institute of Computer Graphics and Vision, Graz University of Technology. pp. 1–40.
- ↑ Wild, Daniel; Weber, Maximilian; Egger, Jan (2019). "Client/Server Based Online Environment for Manual Segmentation of Medical Images". arXiv:1904.08610 [cs.CV].
- ↑ "Studierstube". https://www.cg.tuwien.ac.at/research/vr/studierstube/jvrs-paper.pdf.
- ↑ Szalavári, Zsolt; Schmalstieg, Dieter; Fuhrmann, Anton; Gervautz, Michael (1998). "?Studierstube?: An environment for collaboration in augmented reality" (in en). Virtual Reality. Lecture Notes in Computer Science (Springer International Publishing) 3: 37–48. doi:10.1007/BF01409796.
- ↑ "Python". https://www.python.org/.
- ↑ "The Insight Toolkit". https://itk.org/.
- ↑ "VTK - The Visualization Toolkit". https://vtk.org/.
- ↑ "The X Toolkit: WebGL™ for Scientific Visualization". 25 April 2020. https://github.com/xtk/X.
- ↑ "Slice:Drop". https://slicedrop.com/.
- ↑ "Ajax - Web developer guides". https://developer.mozilla.org/en-US/docs/Web/Guide/AJAX.
- ↑ "1 Scope and Field of Application". http://dicom.nema.org/medical/dicom/current/output/chtml/part01/chapter_1.html#sect_1.1.
- ↑ DICOM brochure, nema.org.
- ↑ Aja-Fernández, Santiago; de Luis Garcia, Rodrigo; Tao, Dacheng; Li, Xuelong (2009). Tensors in Image Processing and Computer Vision. Advances in Computer Vision and Pattern Recognition. Springer Science & Business Media. ISBN 9781848822993.
- ↑ Bhandari, Abhishta; Koppen, Jarrad; Agzarian, Marc (2020). "Convolutional neural networks for brain tumour segmentation" (in en). Insights into Imaging. Lecture Notes in Computer Science (Springer Open) 11:77 (1): 77. doi:10.1186/s13244-020-00869-4. PMID 32514649.
- ↑ Egger, Jan (2017-03-12). "HTC Vive MeVisLab integration via OpenVR for medical applications" (in en-US). PLOS ONE 12 (3): e0173972. doi:10.1371/journal.pone.0173972. PMID 28323840. Bibcode: 2017PLoSO..1273972E.
- ↑ "ParaView". https://www.paraview.org.
External links
 |