Software:Molecular Evolutionary Genetics Analysis
 | |
| Original author(s) | Masatoshi Nei, Sudhir Kumar, Koichiro Tamura, Glen Stecher, Daniel Peterson, Nicholas Peterson |
|---|---|
| Developer(s) | Pennsylvania State University |
| Initial release | 1993 |
| Stable release | 11.0.11
/ February 2022[1] |
| Operating system | Windows, OS X, Linux |
| Platform | x86, x86-64 |
| Available in | English |
| Type | Bioinformatics |
| License | Proprietary freeware |
| Website | www |
Molecular Evolutionary Genetics Analysis (MEGA) is computer software for conducting statistical analysis of molecular evolution and for constructing phylogenetic trees. It includes many sophisticated methods and tools for phylogenomics and phylomedicine. It is licensed as proprietary freeware. The project for developing this software was initiated by the leadership of Masatoshi Nei in his laboratory at the Pennsylvania State University in collaboration with his graduate student Sudhir Kumar and postdoctoral fellow Koichiro Tamura.[2] Nei wrote a monograph (pp. 130) outlining the scope of the software and presenting new statistical methods that were included in MEGA. The entire set of computer programs was written by Kumar and Tamura. The personal computers then lacked the ability to send the monograph and software electronically, so they were delivered by postal mail. From the start, MEGA was intended to be easy-to-use and include solid statistical methods only.
MEGA version 2 (MEGA2), which was coauthored by an additional investigator Ingrid Jakobson, was released in 2001.[3] All the computer programs and the readme files of this version could be sent electronically due to advances in computer technology. Around this time, the leadership of the MEGA project was taken over by Kumar (now at Temple University) and Tamura (now at Tokyo Metropolitan University). The monograph Molecular Evolutionary Genetics Analysis was often used as a textbook for new ways to study molecular evolution.
MEGA has been updated and expanded several times and currently all these versions are available from the MEGA website. The latest release, MEGA7, has been optimized for use on 64-bit computing systems. MEGA is in two version. A graphical user interface is available as a native Microsoft Windows program. A command line version, MEGA-Computing Core (MEGA-CC), is available for native cross-platform operation. The method is widely used and cited. With millions of downloads across the releases, MEGA is cited in more than 85,000 papers. The 5th version has been cited over 25,000 times in 4 years.[4]
Release history
| Version | Release date |
|---|---|
| 1.0 | 1993[2] |
| 1.1 | 1994[6] |
| 2.0 | 2000[3] |
| 2.1 | 2001[3] |
| 3.0 | 2004[7] |
| 4.0 | 2006[8] |
| 4.1 | 2008[8] |
| 5.0 | 2011[9] |
| 5.1 | Oct 2012[9] |
| 5.2 | April 2013[9] |
| 6.0 | December 2013[4] |
| 7.0 | January 2016[10] |
| 10.0 (X) | May 2018[11] |
| 11.0 | February 2022[1] |
Features
Sequence alignment construction
Alignment Editor ― Within MEGA, the Alignment Editor is a tool that may be used for editing and building multiple sequence alignments. The Alignment Editor in MEGA includes an integrated tool for both ClustalW and MUSCLE programs. All actions take place in the Analysis Explorer, which can be found in the main menu of MEGA. When a new alignment is being created, the user is presented with three options: create a new alignment, open a saved alignment session, or retrieve sequences from a file (importing sequences from NCBI). Once an option is selected, the user can choose either ClustalW or MUSCLE from the Alignment tab located at the top of the page. Parameters for the selected alignment program can then be specified and a progress bar will appear while the tool is being computer. Aligned sequences will replace unaligned ones in the main section of the Alignment Editor. To perform further analysis in MEGA, it is advisable to save the alignment session in either MEGA or FASTA format.[12]
Trace Data File Viewer/Editor ― The Trace Data File Viewer/Editor has many functionalities in the following three menus. All the commands are used to help specialize searches and alignments in MEGA.
- Data Menu consists of Open File in New Window, Open File, Save File, Print, Add to Alignment Explore, Export FASTA File, and Exit.[13]
- Edit Menu consists of Undo, Copy, Mask Upstream, Mask Downstream, and Reverse Complement. The difference between Mask Upstream and Mask Downstream is upstream is used to mask/unmask a region to the left of a cursor while downstream does the opposite. Reverse Complement will be used in situations where the complements may need to be reversed in a sequence.[13]
- Search Menu consists of Find, Find Next, Find Previous, Next N, Find in File, and DO BLAST Search. The Find, Find Next, and Find Previous are used to find occurrences in certain sections of a query sequence. Next N is a command the will be able to go to the next indeterminate (N) nucleotide. Find in a File allows a user to search another file for selected sequences. Do BLAST Search command will perform a BLAST search in a separate web browser. The user may be able to either select certain date to BLAST or all sequences in the session will be used.[13]
Integrated web browser, sequence fetching ― MEGA comes with a built-in web browser that allows users to access GenBank sequence data from the NCBI website. The integrated web browser can be accessed when creating a new alignment in the Alignment Editor. To successfully use sequences from NCBI, it is advised to change the searches to FASTA format and use the “Add to Alignment” button. Once completed, all the sequences will be imported into the MEGA application.[14]
Data handling
One of the challenges associated with evolutionary genetic analysis is the presence of ambiguous states such as R, Y, and T. These states often arise from sequence errors or incomplete datasets. However, MEGA offers several resources to handle ambiguous states, including the deletion of sites that have an ambiguity score higher than the Site Coverage Cutoff parameter.[15]
MEGA's extended format allows users to save all data attributes, such as sequence length, nucleotide positions, gaps, and ambiguous states.[16] Additionally, MEGA supports data import from other formats, such as Clustal, which ensures a seamless transition between popular file types.[17]
After importing a dataset, MEGA provides multiple different data viewer options. For example, users can view statistical attributes and select subsets in the Sequence Data Explorer or use the Distance Data Explorer to inspect pairwise distance data.[18] Another feature of MEGA is the visual specification of domain groups. This allows users to group sequences by a specific characteristic and view subsequent phylogenetic trees.
Genetic code table
MEGA offers support for modifying the genetic code used for translating DNA sequences. By default, MEGA has 23 built-in genetic code variations including the standard code, vertebrate mitochondrial code, Drosophila mitochondrial code, and yeast mitochondrial code.[19] Users may add, remove, or edit any genetic code table.
| Genetic code name |
|---|
| Standard |
| Vertebrate Mitochondrial |
| Invertebrate Mitochondrial |
| Yeast Mitochondrial |
| Mold Mitochondrial |
| Protozoan Mitochondrial |
| Coelenterate Mitochondrial |
| Mycoplasma |
| Spiroplasma |
| Ciliate Nuclear |
| Dasycladacean Nuclear |
| Hexamita Nuclear |
| Echinoderm Mitochondrial |
| Euplotid Nuclear |
| Bacterial Plastid |
| Plant Plastid |
| Alternative Yeast Nuclear |
| Ascidian Mitochondrial |
| Flatworm Mitochondrial |
| Blepharisma Mitochondrial |
| Chlorophycean Mitochondrial |
| Trematode Mitochondrial |
| Scenedesmus obliquus Mitochondrial |
| Thraustochytrium Mitochondrial |
In addition, MEGA can also computes the degeneracy of each codon position in a genetic code table as well as the number of synonymous sites and non-synonymous sites using the Nei-Gojobori method.[20]
Real-time caption expert engine
The Caption Expert is a part of MEGA which provides publication-like detailed captions based on the properties of analysis results. It is a tool that may be used for distance matrix, phylogeny, tests, etc. within MEGA (megasoftware).[21]

Figure 1 Above is a phylogenetic tree in MEGA using a Neighbor Joining Method and Bootstrapping. The data sequences being used is from the study Initial diversification of living amphibians predated the breakup of Pangaea. MEGA has created captions using the Real-Time Caption Editor to be able to analyze the properties of the results of the phylogenetic tree. This allows a user to be able to follow and interpret final results.[22]
Integrated text file editor
MEGA's integrated text file editor enables users to edit text files without the need for another program. Features like columnar block selection-editing aid in the performance of bulk operations, like changing letter case or font size. Additionally, the editor includes line numbers to assist with the navigation of large files and identifying areas of interest.[23]
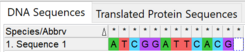
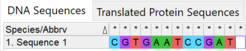
MEGA also provides several tools to format sequences. For example, the built-in reverse complement utility reverses the order of characters and replaces each with its complement.[24]
The screenshots demonstrate the use of MEGA's reverse complement tool. The original sequence was reversed and each nucleotide was replaced with its complement to produce the reverse complement.
Sequence data viewer
MEGA provides a graphical interface for displaying and manipulating aligned nucleotide and protein sequences.[25] The Sequence Data Explorer has multiple menu functionalities to help with exporting data, searching alignments, changing display features, highlighting sites, and computing statistics:
- Data Menu consists of Write Data to File, Translate/Untranslate, Select Genetic Code Table, Setup/Select Genes and Domains, Setup/Select Taxa and Groups, Quit Data Viewer.[26] Translate/Untranslate and Select Genetic Code Table is only available for nucleotide sequences.
- Search Menu consists of Find Motif, Find Next, Find Previous, Find Marked Site, and Highlight Motif.[27]
- Display Menu consists of Show Only Selected Sequences, Use Identical Symbol, Color Cells, Sort Sequences, Restore Input Order, Show Sequence Name, Show Group Name, and Change Font.[28]
- Highlight Menu consists of Conserved Sites, Variable Sites, Parsimony-Informative Sites, Singleton Sites, 0-fold Degenerate Sites, 2-fold Degenerate Sites, and 4-fold Degenerate Sites.[29]
- Statistics Menu consists of Nucleotide Composition, Nucleotide Pair Frequencies, Codon Usage, Amino Acid Composition, Use All Selected Sites, Use only Highlighted Sites, Display Results in Spreadsheet (Excel or Libre/Open Office), Display Results in Comma-Delimited (CSV), Display Result in Text Editor.[30]
MCL-based estimation of nucleotide substitution patterns
Substitution Models in MEGA allow various options with different attributes of substitution models for both DNA and protein sequences. You may choose different substitution types, model, etc. to fit best with chosen data. The three main substitution models are 4x4 Rate Matrix, Transition-Transversion Rate Ratio (k1,k2), and Transition-Transversion Rate Bias of R.
Transition-Transversion Rate Ratios (k1, k2) – Transition-Transversion Rate Ratio calculates the ratio rate of Transition(a) to Transversion(b) using the formula k = a/b.[31]
Transition-Transversion Rate Bias (R) — Transition-Transversion Rate Bias of R in MEGA calculates the ratio of the number of transitions to the number of transversions between a pair of sequences. MEGA allows a user to conduct an analysis of the data with a specified value of R. A key takeaway is when R equals 0.5, it means there is no bias towards either a transition or transversion substitution.[31]
Substitution pattern homogeneity test
MEGA offers several approaches for testing substitution pattern homogeneity, such as composition distance, disparity index, and Monte Carlo tests. These methods are used to determine if different genetic regions evolved under the same selective pressure.
Computation distance measures the variation in nucleotide composition between two sequences. MEGA computes this figure per site and excludes any gaps or missing data. A larger distance suggests that the regions evolved under different selective pressures.[32]
The disparity index evaluates the difference in substitution patterns for a given pair of sequences. This value is calculated per site and is thought to be more dynamic than the chi-square test. A large difference implies that the pattern of substitution was not the same for the given pair of sequences.[32]
The Monte Carlo test is another approach to test substitution pattern homogeneity that involves running a null distribution simulation. MEGA requires the user to specify the number of replicates and a starting seed. For a significant result, many simulations must be performed.[32] Therefore, it is essential to consider the computational cost of the algorithm.
| Monte Carlo method | Computational complexity |
|---|---|
| MC + exact simulation | |
| MC + tau-leaping | |
| MC + midpt. or trap. tau-leaping | |
| MC + Euler for diff. approx. |
The table above shows the computational complexity of different Monte Carlo methods as approaches infinity in relation to the parameter . While it's not clear which method MEGA employs, it is likely to be computationally intensive because all the methods listed in the table have a computational order greater than .
Distance estimation methods
MEGA offers a wide variety of options for calculating evolutionary distance between a pair of nucleotide or amino acid sequences with or without standard errors.[34] Distance methods are divide into three categories, nucleotide, syn-nonsynonymous, and amino acids:
- Nucleotide - Sequences are compared nucleotide-by-nucleotide, available for both protein coding and non-coding sequences.[34]
- No. of differences
- p-distance
- Juke-Cantor Model
- Tajima-Nei Model
- Kimura 2-Parameter Model
- Tamura 3-Parameter Model
- Tamura-Nei Model
- Log-Det Method
- Maximum Composite Likelihood Model
- Syn-Nonsynonymous - Sequences are compared codon-by-codon, only available for protein coding sequences.[35]
- Nei-Gojobori Method
- Modified Nei-Gojobori Method
- Li-Wu-Luo Method
- Pamilo-Bianchi-Li Method
- Kumar Method
- Amino Acid - Sequences are compared residue-by-residue, available for both protein coding and non-coding sequences.[36]
- No. of differences
- p-distance
- Poisson Model
- Equal Input Model
- Dayhoff Model
- Jones-Taylor-Thornton Model
After selecting a distance method, a subset of attributes will become visible when applicable. The attributes are Substitutions to Include, Transition/Transversion Ratio, Pattern among Lineages, and Rates among Sites. For example, if a model has a rate variation, the gamma parameter will become visible. In addition, every distance method provides options for handling gap and missing data, and codon position if applicable.[37]
Every substitution matrix has it own use case. One of the simplest model is the Juke-Cantor, which assumes an equal mutation rates. The Kimura 2-Parameter model extends that model but with distinctions between transition rates ( and ) and transversion rates (). Then the Kimura 3-Parameter model extends that model but with distinctions between transversions that conserve the nucleotide's weak/strong property ( and ) and transversions that conserve the nucleotide's amino/keto property ( and ). However, each extension adds more parameter and risk the issue of overfitting. The best substitution matrix depends on the data used. To help with selection, MEGA provides a Find Best-Fit Substitution Model in the Model tab that run each model and assigns a Bayesian information criterion evaluation.
Tests of selection
Large sample Z-test The Z-test is used to compare relative synonymous and nonsynonymous substitutions within a gene sequence, with the main objective of determining positive selection. To perform the Z-test formula, an estimation of the number of synonymous substitutions per synonymous site (dS) and nonsynonymous substitutions per nonsynonymous site (dN) must be account for, along with the variances of the synonymous and nonsynonymous substitutions Var(dS) and Var(dN). The formula used for the Z-test is:
Z = (dN – dS_ / SQRT(Var(dS) + Var(dN))
If dN is greater than dS, it indicates positive selection, while if dN is less than dS, it indicates purifying selection. The output of Z from the formula above will determine if it is a positive or purifying selection. Key factors to determine which selection the output will be is the variances of the synonymous and nonsynonymous sites. These tests are commonly used for analytical formulas or bootstrapping resampling in MEGA.[38]
Fisher's exact test — Fisher's Exact Test examines synonymous and nonsynonymous substitutions in sequences and is referred to as a one-tailed test when analyzing small samples for positive selection. Rejecting the null hypothesis of neutrality occurs when the P-Value is less than 0.05. If the differences per synonymous site exceed those per nonsynonymous site, MEGA assigns a P-Value of 1, indicating purifying selection rather than positive selection.[39] Further research on Fisher's Exact Test, the algorithm is based on the probability distribution of n!. As a conclusion, it could be argued that the time complexity of the algorithm is O(n!). The name for the distribution method is Hypergeometric Distribution (Hoffman).[40]
Tajima's Neutrality Test — The purpose of Tajima's Neutrality Test is to assess the relationship between the number of segregating sites per site and nucleotide diversity. When alleles are selectively neutral the product 4Nv can be estimated in two ways. N represents the effective population size and v is the mutation rate per site. By calculating the difference between these estimates, one can determine if there is evidence of non-neutral evolution.[41]
Molecular clock test
The molecular clock hypothesis suggests that all sequences have evolved at a constant rate over time. Therefore, the molecular clock test evaluates this statement in conjunction with the data provided by the user. In MEGA, this test is performed by applying a maximum likelihood test to a given tree topology and sequence alignment. This produces two log-likelihood values, one with the clock hypothesis and one without.[42]
Another approach offered by MEGA is Tajima's relative rate test. This method compares the number of substitutions per site between different sequences. If the resulting numbers differ by a large factor, the molecular clock hypothesis may not be valid for the given data set.[43]
Tree-making methods
MEGA offers five methods building a phylogenetic tree:
- Neighbor joining
- Minimum evolution
- UPGMA
- Maximum parsimony
- Maximum likelihood
Each method allows for a bootstrap phylogeny test with any number of replications. Neighbor joining and minimum evolution allows for an interior-branch test instead. Substitution model and parameters are the same as the distance estimation methods.
Tree explorers


MEGA provides a graphical interface for displaying a phylogenetic tree based on a variety of options. In the view menu, the tree can be displayed in three different styles: traditional, radiation, or circle. Traditional trees have three different branch styles: rectangular, straight, or curved. The view menu also offers toggling topology scaling, changing font type and size, arranging taxa, showing/hiding various details, and a general option for more control over the tree drawing aspects.[44]
The subtree menu provides options for manipulating the tree, such as swapping branches, flipping lineages order, compressing/expanding subtrees, and moving the tree's root. Subtrees can also be displayed in its own tree explorer with all the same features and options.[45] The compute menu provides options for computing a condensed tree, a consensus tree, or a timetree with or without a molecular clock.[46] The file menu provides options for saving, exporting, printing, and exiting. The tree topology can be exported to a file in MEGA tree format, or for timetrees, exported in a tabular format with relevant information used when constructing the timetree. Other export options include the current timetree calibrations, analysis summary, partition list, and pairwise distances.[47] The tree explorer also provide options to save the current tree display in an image format or to the clipboard under the image menu option. The image format supported are BMP, PNG, PDF, SVG, TIFF, and EMF.[48] If the user chose to build the tree with bootstrap replication, then the tree explorer will have two tabs, one with the original tree and one with the bootstrap consensus tree.
External links
References
- ↑ 1.0 1.1 "Update History". https://www.megasoftware.net/history.
- ↑ 2.0 2.1 Kumar, S., K. Tamura, and M. Nei (1993) MEGA: Molecular Evolutionary Genetics Analysis. Ver. 1.0, The Pennsylvania State University, University Park, PA.
- ↑ 3.0 3.1 3.2 Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei (2001) MEGA2: Molecular Evolutionary Genetics Analysis. Ver. 2.0, Bioinformatics 17:1244-1245.
- ↑ 4.0 4.1 Tamura, K., Stecher, G., Peterson, D., Filipski, A., Kumar, S. (2013) MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30:2725-2729.
- ↑ "MEGA Update History". http://www.megasoftware.net/history.
- ↑ Kumar, S., K. Tamura, and M. Nei (1994) MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. CABIOS 10:189-191.
- ↑ Kumar, S., Tamura, K., and Nei, M. (2004) MEGA3: Molecular Evolutionary Genetics Analysis. Brief. Bioinformatics 5:150-163.
- ↑ 8.0 8.1 Tamura, K., Dudley, J., Nei, M., Kumar, S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis. Mol. Biol. Evol. 24(8):1596-1599.
- ↑ 9.0 9.1 9.2 Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., Kumar, S. (2011) MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 28:2731-2739.
- ↑ Kumar, S., Stecher, G., & Tamura, K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33(7):1870–1874.
- ↑ Kumar, S., Stecher, G., Li M., Knyaz C. & Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 35(6):1547-1549.
- ↑ "Aligning Sequences Manual Page". https://www.megasoftware.net/web_help_11/Part_I_Getting_Started/A_Walk_Through_MEGA/Aligning_Sequences.htm.
- ↑ 13.0 13.1 13.2 "Trace Data File Viewer/Editor". https://www.megasoftware.net/web_help_11/Trace_Data_File_Viewer_Editor.htm.
- ↑ "Obtaining Sequence Data from the Internet (GenBank)". https://www.megasoftware.net/web_help_11/index.htm#t=Part_I_Getting_Started%2FA_Walk_Through_MEGA%2FAligning_Sequences.htm&rhsyns=%20&rhsearch=Integrated%20web%20browser%2C%20sequence%20fetching&rhhlterm=Integrated%20web%20browser%2C%20sequence%20fetchingn.
- ↑ "Partial Deletion Manual Page". https://www.megasoftware.net/web_help_11/index.htm#t=Partial-Deletion_Option.htm&rhsearch=ambiguous&rhhlterm=ambiguous&rhsyns=%20.
- ↑ "Saved Sessions Manual Page". https://www.megasoftware.net/web_help_11/index.htm#t=Part_III_Input_Data_Types_and_File_Format%2FMEGA_Input_Data_File_Formats%2FMEGA_Saved_Sessions.htm&rhsearch=save&rhsyns=%20.
- ↑ "Import Data Manual Page". https://www.megasoftware.net/web_help_11/index.htm#t=Importing_Data.htm&rhsearch=import&rhsyns=%20&rhhlterm=import.
- ↑ "Data Explorer Manual Page". https://www.megasoftware.net/web_help_11/index.htm#t=Data_Explorer.htm&rhsearch=Data%20Explorer&rhsyns=%20&rhhlterm=Data%20Explorer.
- ↑ "Built-in Genetic Codes". https://www.megasoftware.net/web_help_11/Built-in_Genetic_Codes.htm.
- ↑ "Genetic Code Statistical Attributes". https://www.megasoftware.net/web_help_11/Genetic_Code_Statistical_Attributes.htm.
- ↑ "Creating Data Captions with Caption Expert". https://www.megasoftware.net/mega4/WebHelp/hc_caption_expert.htm.
- ↑ San Mauro, Diego; Vences, Miguel; Alcobendas, Mariana (17 March 2005). "Initial diversification of living amphibians predated the breakup of Pangaea.". https://www.ncbi.nlm.nih.gov/.
- ↑ "Text Editor Manual Page". https://www.megasoftware.net/web_help_11/index.htm#t=Part_II_Assembling_Data_For_Analysis%2FText_Editor_And_Utilities%2FText_Editor.htm&rhsearch=text%20file%20editor&rhhlterm=text%20file%20editor&rhsyns=%20.
- ↑ "Reverse Complement Manual Page". https://www.megasoftware.net/web_help_11/index.htm#t=Part_II_Assembling_Data_For_Analysis%2FText_Editor_And_Utilities%2FReverse_Complement.htm&rhsearch=%20reverse%20complement&rhhlterm=%20reverse%20complement&rhsyns=%20.
- ↑ "Sequence Data Explorer". https://www.megasoftware.net/web_help_11/Sequence_Data_Explorer.htm.
- ↑ "Data Menu (Sequence Data Explorer)". https://www.megasoftware.net/web_help_11/Data_Menu_(Sequence_Data_Explorer).htm.
- ↑ "Search Menu". https://www.megasoftware.net/web_help_11/Part_II_Assembling_Data_For_Analysis/Building_Sequence_Alignments/Menu_Items_In_Alignment_Explorer/Search_Menu.htm.
- ↑ "Display Menu (in Sequence Data Explorer)". https://www.megasoftware.net/web_help_11/Display_Menu_in_Sequence_Data_Explorer.htm.
- ↑ "Highlight Menu (in Sequence Data Explorer)". https://www.megasoftware.net/web_help_11/Highlight_Menu_in_Sequence_Data_Explorer.htm.
- ↑ "Statistics Menu (in Sequence Data Explorer)". https://www.megasoftware.net/web_help_11/Statistics_Menu_in_Sequence_Data_Explorer.htm.
- ↑ 31.0 31.1 "Transition/Transversion Ratio (R)". https://www.megasoftware.net/web_help_7/rh_transition_transversion_ratio_r.htm.
- ↑ 32.0 32.1 32.2 "Pattern Homogeneity Analysis Manual Page". https://www.megasoftware.net/web_help_11/index.htm#t=Analysis_Preferences_Pattern_Homogeneity_Analysis.htm&rhsearch=Substitution%20pattern%20homogeneity&rhsyns=%20&rhhlterm=Substitution%20pattern%20homogeneity.
- ↑ Anderson, David F.; Higham, Desmond J.; Sun, Yu (2018). "Computational Complexity Analysis for Monte Carlo Approximations of Classically Scaled Population Processes". Multiscale Modeling & Simulation 16 (3): 1206–1226. doi:10.1137/17M1138169. https://doi.org/10.1137/17M1138169.
- ↑ 34.0 34.1 "Models for estimating distances". https://www.megasoftware.net/web_help_11/Models_for_estimating_distances.htm.
- ↑ "Syn-Nonsynonymous". https://www.megasoftware.net/web_help_11/Syn-Nonsynonymous.htm.
- ↑ "Amino Acid". https://www.megasoftware.net/web_help_11/Amino_Acid.htm.
- ↑ "Analysis Preferences (Distance Computation)". https://www.megasoftware.net/web_help_11/Analysis_Preferences_Distance_Computation.htm.
- ↑ "Codon Based Z-Test (large sample)". https://www.megasoftware.net/web_help_11/index.htm#t=Codon_Based_Z-Test_large_sample.htm&rhsyns=%20&rhsearch=large%20sample%20z%20test&rhhlterm=large%20sample%20z%20test.
- ↑ "Codon Based Fisher's Exact Test". https://www.megasoftware.net/web_help_11/index.htm#t=Codon_Based_Fisher_s_Exact_Test.htm&rhsyns=%20&rhsearch=-%09Fisher%E2%80%99s%20exact%20test&rhhlterm=-%09Fisher%E2%80%99s%20exact%20test.
- ↑ I.E. Hoffman, Julien (2015). "Hypergeometric Distribution". Elsevier Inc.. https://www.sciencedirect.com/topics/medicine-and-dentistry/fisher-exact-test.
- ↑ "Tajima's Test of Neutrality". https://www.megasoftware.net/web_help_11/index.htm#t=Tajima_s_Test_of_Neutrality.htm&rhsyns=%20&rhsearch=Tajima%E2%80%99s%20neutrality%20test%20&rhhlterm=Tajima%E2%80%99s%20neutrality%20test%20.
- ↑ "Molecular Clock Test Manual Page". https://www.megasoftware.net/web_help_11/index.htm#t=Molecular_Clock_Test_ML.htm&rhsearch=molecular%20clock&rhsyns=%20&rhhlterm=molecular%20clock.
- ↑ "Tajima's Test Manual Page". https://www.megasoftware.net/web_help_11/index.htm#t=Tajima_s_Test_Relative_Rate.htm&rhsearch=Tajima%E2%80%99s&rhsyns=%20&rhhlterm=Tajima%E2%80%99s.
- ↑ "View Menu (in Tree Explorer)". https://www.megasoftware.net/web_help_11/View_Menu_in_Tree_Explorer.htm.
- ↑ "Subtree Menu (in Tree Explorer)". https://www.megasoftware.net/web_help_11/Subtree_Menu_in_Tree_Explorer.htm.
- ↑ "Calibrate Timetree with Molecular Clock". https://www.megasoftware.net/web_help_11/Calibrate_Timetree_with_Molecular_Clock.htm.
- ↑ "File Menu (in Tree Explorer)". https://www.megasoftware.net/web_help_11/TreeExplorer_HC_files/File_Menu_in_Tree_Explorer.htm.
- ↑ "Image Menu (in Tree Explorer)". https://www.megasoftware.net/web_help_11/Image_Menu_in_Tree_Explorer.htm.
