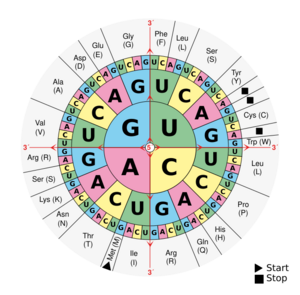
Biology:DNA and RNA codon tables
A codon table can be used to translate a genetic code into a sequence of amino acids.[1][2] The standard genetic code is traditionally represented as an RNA codon table, because when proteins are made in a cell by ribosomes, it is messenger RNA (mRNA) that directs protein synthesis.[2][3] The mRNA sequence is determined by the sequence of genomic DNA.[4] In this context, the standard genetic code is referred to as translation table 1.[3] It can also be represented in a DNA codon table. The DNA codons in such tables occur on the sense DNA strand and are arranged in a 5′-to-3′ direction. Different tables with alternate codons are used depending on the source of the genetic code, such as from a cell nucleus, mitochondrion, plastid, or hydrogenosome.[5]
There are 64 different codons in the genetic code and the below tables; most specify an amino acid.[6] Three sequences, UAG, UGA, and UAA, known as stop codons,[note 1] do not code for an amino acid but instead signal the release of the nascent polypeptide from the ribosome.[7] In the standard code, the sequence AUG—read as methionine—can serve as a start codon and, along with sequences such as an initiation factor, initiates translation.[3][8][9] In rare instances, start codons in the standard code may also include GUG or UUG; these codons normally represent valine and leucine, respectively, but as start codons they are translated as methionine or formylmethionine.[3][9]
The first table—the standard table—can be used to translate nucleotide triplets into the corresponding amino acid or appropriate signal if it is a start or stop codon. The second table, appropriately called the inverse, does the opposite: it can be used to deduce a possible triplet code if the amino acid is known. As multiple codons can code for the same amino acid, the International Union of Pure and Applied Chemistry's (IUPAC) nucleic acid notation is given in some instances.
Translation table 1
Standard RNA codon table
| Amino-acid biochemical properties | Nonpolar (np) | Polar (p) | Basic (b) | Acidic (a) | Termination: stop codon * | Initiation: possible start codon ⇒ |
| 1st base |
2nd base | 3rd base | |||||||
|---|---|---|---|---|---|---|---|---|---|
| U | C | A | G | ||||||
| U | UUU | (Phe/F) Phenylalanine (np) | UCU | (Ser/S) Serine (p) | UAU | (Tyr/Y) Tyrosine (p) | UGU | (Cys/C) Cysteine (p) | U |
| UUC | UCC | UAC | UGC | C | |||||
| UUA | (Leu/L) Leucine (np) | UCA | UAA | Stop (Ochre) *[note 2] | UGA | Stop (Opal) *[note 2] | A | ||
| UUG ⇒ | UCG | UAG | Stop (Amber) *[note 2] | UGG | (Trp/W) Tryptophan (np) | G | |||
| C | CUU | CCU | (Pro/P) Proline (np) | CAU | (His/H) Histidine (b) | CGU | (Arg/R) Arginine (b) | U | |
| CUC | CCC | CAC | CGC | C | |||||
| CUA | CCA | CAA | (Gln/Q) Glutamine (p) | CGA | A | ||||
| CUG | CCG | CAG | CGG | G | |||||
| A | AUU | (Ile/I) Isoleucine (np) | ACU | (Thr/T) Threonine (p) | AAU | (Asn/N) Asparagine (p) | AGU | (Ser/S) Serine (p) | U |
| AUC | ACC | AAC | AGC | C | |||||
| AUA | ACA | AAA | (Lys/K) Lysine (b) | AGA | (Arg/R) Arginine (b) | A | |||
| AUG ⇒ | (Met/M) Methionine (np) | ACG | AAG | AGG | G | ||||
| G | GUU | (Val/V) Valine (np) | GCU | (Ala/A) Alanine (np) | GAU | (Asp/D) Aspartic acid (a) | GGU | (Gly/G) Glycine (np) | U |
| GUC | GCC | GAC | GGC | C | |||||
| GUA | GCA | GAA | (Glu/E) Glutamic acid (a) | GGA | A | ||||
| GUG ⇒ | GCG | GAG | GGG | G | |||||
As shown in the above table, NCBI table 1 includes the less-canonical start codons GUG and UUG.[3]
Inverse RNA codon table
| Amino acid | RNA codons | Compressed | Amino acid | RNA codons | Compressed | |
|---|---|---|---|---|---|---|
| Ala, A | GCU, GCC, GCA, GCG | GCN | Ile, I | AUU, AUC, AUA | AUH | |
| Arg, R | CGU, CGC, CGA, CGG; AGA, AGG | CGN, AGR; or CGY, MGR |
Leu, L | CUU, CUC, CUA, CUG; UUA, UUG | CUN, UUR; or CUY, YUR | |
| Asn, N | AAU, AAC | AAY | Lys, K | AAA, AAG | AAR | |
| Asp, D | GAU, GAC | GAY | Met, M | AUG | ||
| Asn or Asp, B | AAU, AAC; GAU, GAC | RAY | Phe, F | UUU, UUC | UUY | |
| Cys, C | UGU, UGC | UGY | Pro, P | CCU, CCC, CCA, CCG | CCN | |
| Gln, Q | CAA, CAG | CAR | Ser, S | UCU, UCC, UCA, UCG; AGU, AGC | UCN, AGY | |
| Glu, E | GAA, GAG | GAR | Thr, T | ACU, ACC, ACA, ACG | ACN | |
| Gln or Glu, Z | CAA, CAG; GAA, GAG | SAR | Trp, W | UGG | ||
| Gly, G | GGU, GGC, GGA, GGG | GGN | Tyr, Y | UAU, UAC | UAY | |
| His, H | CAU, CAC | CAY | Val, V | GUU, GUC, GUA, GUG | GUN | |
| START | AUG, CUG, UUG | HUG | STOP | UAA, UGA, UAG | URA, UAG; or UGA, UAR | |
Standard DNA codon table
| Amino-acid biochemical properties | Nonpolar (np) | Polar (p) | Basic (b) | Acidic (a) | Termination: stop codon * | Initiation: possible start codon ⇒ |
| 1st base |
2nd base | 3rd base | |||||||
|---|---|---|---|---|---|---|---|---|---|
| T | C | A | G | ||||||
| T | TTT | (Phe/F) Phenylalanine (np) | TCT | (Ser/S) Serine (p) | TAT | (Tyr/Y) Tyrosine (p) | TGT | (Cys/C) Cysteine (p) | |
| TTC | C | ||||||||
| TTA | (Leu/L) Leucine (np) | Stop (Ochre) *[note 2] | Stop (Opal) *[note 2] | A | |||||
| TTG ⇒ | Stop (Amber) *[note 2] | (Trp/W) Tryptophan (np) | G | ||||||
| C | CTT | (Pro/P) Proline (np) | (His/H) Histidine (b) | (Arg/R) Arginine (b) | |||||
| CCC | CAC | CGC | C | ||||||
| CCA | CAA | (Gln/Q) Glutamine (p) | CGA | A | |||||
| CCG | CAG | CGG | G | ||||||
| A | ATT | (Ile/I) Isoleucine (np) | (Thr/T) Threonine (p) | (Asn/N) Asparagine (p) | (Ser/S) Serine (p) | ||||
| ACC | AAC | AGC | C | ||||||
| ACA | AAA | (Lys/K) Lysine (b) | AGA | (Arg/R) Arginine (b) | A | ||||
| ATG ⇒ | (Met/M) Methionine (np) | ACG | AAG | AGG | G | ||||
| G | GTT | (Val/V) Valine (np) | (Ala/A) Alanine (np) | (Asp/D) Aspartic acid (a) | (Gly/G) Glycine (np) | ||||
| GCC | GAC | GGC | C | ||||||
| GCA | GAA | (Glu/E) Glutamic acid (a) | GGA | A | |||||
| GTG ⇒ | GCG | GAG | GGG | G | |||||
Inverse DNA codon table
| Amino acid | DNA codons | Compressed | Amino acid | DNA codons | Compressed | |
|---|---|---|---|---|---|---|
| Ala, A | GCT, GCC, GCA, GCG | GCN | Ile, I | ATT, ATC, ATA | ||
| Arg, R | CGT, CGC, CGA, CGG; AGA, AGG | CGN, AGR; or CGY, MGR |
Leu, L | CTT, CTC, CTA, CTG; TTA, TTG | CTN, TTR; or CTY, YTR | |
| Asn, N | AAT, AAC | AAY | Lys, K | AAA, AAG | AAR | |
| Asp, D | GAT, GAC | GAY | Met, M | |||
| Asn or Asp, B | AAT, AAC; GAT, GAC | RAY | Phe, F | TTT, TTC | TTY | |
| Cys, C | TGT, TGC | Pro, P | CCT, CCC, CCA, CCG | CCN | ||
| Gln, Q | CAA, CAG | CAR | Ser, S | TCT, TCC, TCA, TCG; AGT, AGC | TCN, AGY | |
| Glu, E | GAA, GAG | GAR | Thr, T | ACT, ACC, ACA, ACG | ACN | |
| Gln or Glu, Z | CAA, CAG; GAA, GAG | SAR | Trp, W | |||
| Gly, G | GGT, GGC, GGA, GGG | GGN | Tyr, Y | TAT, TAC | ||
| His, H | CAT, CAC | CAY | Val, V | GTT, GTC, GTA, GTG | ||
| START | ATG, CTG, UTG | HTG | STOP | TAA, TGA, TAG | TRA, TAR |
Alternative codons in other translation tables
This section is missing information about start codon in these tables, aka the "sncbieaa" row in NCBI data. (December 2023) |
The genetic code was once believed to be universal:[16] a codon would code for the same amino acid regardless of the organism or source. However, it is now agreed that the genetic code evolves,[17] resulting in discrepancies in how a codon is translated depending on the genetic source.[16][17] For example, in 1981, it was discovered that the use of codons AUA, UGA, AGA and AGG by the coding system in mammalian mitochondria differed from the universal code.[16] Stop codons can also be affected: in ciliated protozoa, the universal stop codons UAA and UAG code for glutamine.[17][note 4] The following table displays these alternative codons.
| Amino-acid biochemical properties | Nonpolar (np) | Polar (p) | Basic (b) | Acidic (a) | Termination: stop codon * |
| Code | Translation table |
DNA codon involved | RNA codon involved | Translation with this code |
Standard translation | Notes | ||
|---|---|---|---|---|---|---|---|---|
| Standard | 1 | Includes translation table 8 (plant chloroplasts). | ||||||
| Vertebrate mitochondrial | 2 | AGA | AGA | Stop * | Arg (R) (b) | |||
| AGG | AGG | Stop * | Arg (R) (b) | |||||
| ATA | AUA | Met (M) (np) | Ile (I) (np) | |||||
| TGA | UGA | Trp (W) (np) | Stop * | |||||
| Yeast mitochondrial | 3 | ATA | AUA | Met (M) (np) | Ile (I) (np) | |||
| CTT | CUU | Thr (T) (p) | Leu (L) (np) | |||||
| CTC | CUC | Thr (T) (p) | Leu (L) (np) | |||||
| CTA | CUA | Thr (T) (p) | Leu (L) (np) | |||||
| CTG | CUG | Thr (T) (p) | Leu (L) (np) | |||||
| TGA | UGA | Trp (W) (np) | Stop * | |||||
| CGA | CGA | absent | Arg (R) (b) | |||||
| CGC | CGC | absent | Arg (R) (b) | |||||
| Mold, protozoan, and coelenterate mitochondrial + Mycoplasma / Spiroplasma | 4 | TGA | UGA | Trp (W) (np) | Stop * | Includes the translation table 7 (kinetoplasts). | ||
| Invertebrate mitochondrial | 5 | AGA | AGA | Ser (S) (p) | Arg (R) (b) | |||
| AGG | AGG | Ser (S) (p) | Arg (R) (b) | |||||
| ATA | AUA | Met (M) (np) | Ile (I) (np) | |||||
| TGA | UGA | Trp (W) (np) | Stop * | |||||
| Ciliate, dasycladacean and Hexamita nuclear | 6 | TAA | UAA | Gln (Q) (p) | Stop * | |||
| TAG | UAG | Gln (Q) (p) | Stop * | |||||
| Echinoderm and flatworm mitochondrial | 9 | AAA | AAA | Asn (N) (p) | Lys (K) (b) | |||
| AGA | AGA | Ser (S) (p) | Arg (R) (b) | |||||
| AGG | AGG | Ser (S) (p) | Arg (R) (b) | |||||
| TGA | UGA | Trp (W) (np) | Stop * | |||||
| Euplotid nuclear | 10 | TGA | UGA | Cys (C) (p) | Stop * | |||
| Bacterial, archaeal and plant plastid | 11 | See translation table 1. | ||||||
| Alternative yeast nuclear | 12 | CTG | CUG | Ser (S) (p) | Leu (L) (np) | |||
| Ascidian mitochondrial | 13 | AGA | AGA | Gly (G) (np) | Arg (R) (b) | |||
| AGG | AGG | Gly (G) (np) | Arg (R) (b) | |||||
| ATA | AUA | Met (M) (np) | Ile (I) (np) | |||||
| TGA | UGA | Trp (W) (np) | Stop * | |||||
| Alternative flatworm mitochondrial | 14 | AAA | AAA | Asn (N) (p) | Lys (K) (b) | |||
| AGA | AGA | Ser (S) (p) | Arg (R) (b) | |||||
| AGG | AGG | Ser (S) (p) | Arg (R) (b) | |||||
| TAA | UAA | Tyr (Y) (p) | Stop * | |||||
| TGA | UGA | Trp (W) (np) | Stop * | |||||
| Blepharisma nuclear | 15 | TAG | UAG | Gln (Q) (p) | Stop * | As of Nov. 18, 2016: absent from the NCBI update. Similar to translation table 6. | ||
| Chlorophycean mitochondrial | 16 | TAG | UAG | Leu (L) (np) | Stop * | |||
| Trematode mitochondrial | 21 | TGA | UGA | Trp (W) (np) | Stop * | |||
| ATA | AUA | Met (M) (np) | Ile (I) (np) | |||||
| AGA | AGA | Ser (S) | Arg (R) (b) | |||||
| AGG | AGG | Ser (S) (p) | Arg (R) (b) | |||||
| AAA | AAA | Asn (N) (p) | Lys (K) (b) | |||||
| Scenedesmus obliquus mitochondrial | 22 | TCA | UCA | Stop * | Ser (S) (p) | |||
| TAG | UAG | Leu (L) (np) | Stop * | |||||
| Thraustochytrium mitochondrial | 23 | TTA | UUA | Stop * | Leu (L) (np) | Similar to translation table 11. | ||
| Pterobranchia mitochondrial | 24 | AGA | AGA | Ser (S) (p) | Arg (R) (b) | |||
| AGG | AGG | Lys (K) (b) | Arg (R) (b) | |||||
| TGA | UGA | Trp (W) (np) | Stop * | |||||
| Candidate division SR1 and Gracilibacteria | 25 | TGA | UGA | Gly (G) (np) | Stop * | |||
| Pachysolen tannophilus nuclear | 26 | CTG | CUG | Ala (A) (np) | Leu (L) (np) | |||
| Karyorelict nuclear | 27 | TAA | UAA | Gln (Q) (p) | Stop * | |||
| TAG | UAG | Gln (Q) (p) | Stop * | |||||
| TG | UGA | Stop * | or | Trp (W) (np) | Stop * | |||
| Condylostoma nuclear | 28 | TAA | UAA | Stop * | or | Gln (Q) (p) | Stop * | |
| TAG | UAG | Stop * | or | Gln (Q) (p) | Stop * | |||
| TGA | UGA | Stop * | or | Trp (W) (np) | Stop * | |||
| Mesodinium nuclear | 29 | TAA | UAA | Tyr (Y) (p) | Stop * | |||
| TAG | UAG | Tyr (Y) (p) | Stop * | |||||
| Peritrich nuclear | 30 | TA | UAA | Glu (E) (a) | Stop * | |||
| TAG | UAG | Glu (E) (a) | Stop * | |||||
| Blastocrithidia nuclear | 31 | TAA | UAA | Stop * | or | Glu (E) (a) | Stop * | |
| TAG | UAG | Stop * | or | Glu (E) (a) | Stop * | |||
| TGA | UGA | Trp (W) (np) | Stop * | |||||
| Cephalodiscidae mitochondrial code | 33 | AGA | AGA | Ser (S) (p) | Arg (R) (b) | Similar to translation table 24. | ||
| AGG | AGG | Lys (K) (b) | Arg (R) (b) | |||||
| TAA | UAA | Tyr (Y) (p) | Stop * | |||||
| TGA | UGA | Trp (W) (np) | Stop * | |||||
See also
Notes
- ↑ Each stop codon has a specific name: UAG is amber, UGA is opal or umber, and UAA is ochre.[7] In DNA, these stop codons are TAG, TGA, and TAA, respectively.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 The historical basis for designating the stop codons as amber, ochre and opal is described in the autobiography of Sydney Brenner[11] and in a historical article by Bob Edgar.[12]
- ↑ The major difference between DNA and RNA is that thymine (T) is only found in the former. In RNA, it is replaced with uracil (U).[15] This is the only difference between the standard RNA codon table and the standard DNA codon table.
- ↑ Euplotes octacarinatus is an exception.[17]
References
- ↑ 1.0 1.1 "Amino Acid Translation Table". Oregon State University. http://sites.science.oregonstate.edu/genbio/otherresources/aminoacidtranslation.htm.
- ↑ 2.0 2.1 Bartee, Lisa; Brook, Jack. MHCC Biology 112: Biology for Health Professions. Open Oregon. p. 42. https://mhccbiology112.pressbooks.com. Retrieved 6 December 2020.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 "The Genetic Codes". National Center for Biotechnology Information. 7 January 2019. https://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi.
- ↑ "RNA Functions". Nature Education. https://www.nature.com/scitable/topicpage/rna-functions-352/.
- ↑ "The Genetic Codes". National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi.
- ↑ "Codon". https://www.genome.gov/genetics-glossary/Codon.
- ↑ 7.0 7.1 Maloy S. (29 November 2003). "How nonsense mutations got their names". Microbial Genetics Course. San Diego State University. http://www.sci.sdsu.edu/~smaloy/MicrobialGenetics/topics/rev-sup/amber-name.html.
- ↑ "Molecular Mechanism of Scanning and Start Codon Selection in Eukaryotes". Microbiology and Molecular Biology Reviews 75 (3): 434–467. 2011. doi:10.1128/MMBR.00008-11. PMID 21885680.
- ↑ 9.0 9.1 "Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons". Biology of the Cell 95 (3–4): 169–78. 2003. doi:10.1016/S0248-4900(03)00033-9. PMID 12867081.
- ↑ "The Information in DNA Determines Cellular Function via Translation". Nature Education. https://www.nature.com/scitable/topicpage/the-information-in-dna-determines-cellular-function-6523228/.
- ↑ Brenner, Sydney; Wolpert, Lewis (2001). A Life in Science. Biomed Central Limited. pp. 101–104. ISBN 9780954027803.
- ↑ "The genome of bacteriophage T4: an archeological dig". Genetics 168 (2): 575–82. 2004. doi:10.1093/genetics/168.2.575. PMID 15514035. see pages 580–581
- ↑ 13.0 13.1 IUPAC—IUB Commission on Biochemical Nomenclature. "Abbreviations and Symbols for Nucleic Acids, Polynucleotides and Their Constituents". International Union of Pure and Applied Chemistry. http://publications.iupac.org/pac/1974/pdf/4003x0277.pdf.
- ↑ "What does DNA do?". Welcome Genome Campus. https://www.yourgenome.org/facts/what-does-dna-do.
- ↑ "Genes". Boston University. https://sphweb.bumc.bu.edu/otlt/MPH-Modules/PH/DNA-Genetics/DNA-Genetics3.html.
- ↑ 16.0 16.1 16.2 Osawa, A (November 1993). "Evolutionary changes in the genetic code". Comparative Biochemistry and Physiology 106 (2): 489–94. doi:10.1016/0305-0491(93)90122-l. PMID 8281749. https://pubmed.ncbi.nlm.nih.gov/8281749/.
- ↑ 17.0 17.1 17.2 17.3 "Recent evidence for evolution of the genetic code". Microbiological Reviews 56 (1): 229–64. March 1992. doi:10.1128/MR.56.1.229-264.1992. PMID 1579111.
Further reading
- "Case for the genetic code as a triplet of triplets". Proceedings of the National Academy of Sciences of the United States of America 114 (18): 4745–4750. 2 May 2017. doi:10.1073/pnas.1614896114. PMID 28416671. Bibcode: 2017PNAS..114.4745C.
- "A New Start for Protein Synthesis". Science (American Association for the Advancement of Science) 336 (6089): 1645–1646. 29 June 2012. doi:10.1126/science.1224439. PMID 22745408. Bibcode: 2012Sci...336.1645D. https://zenodo.org/record/1230920. Retrieved 17 October 2020.
- "Synthetic polynucleotides and the amino acid code. VII". Proceedings of the National Academy of Sciences of the United States of America 48 (12): 2087–2094. December 1962. doi:10.1073/pnas.48.12.2087. PMID 13946552. Bibcode: 1962PNAS...48.2087G.
- "Evolution and the universality of the mechanism of initiation of protein synthesis". Gene 432 (1–2): 1–6. March 2009. doi:10.1016/j.gene.2008.11.001. PMID 19056476.
- "Synthetic polynucleotides and the amino acid code. VIII". Proceedings of the National Academy of Sciences of the United States of America 49 (1): 116–122. January 1963. doi:10.1073/pnas.49.1.116. PMID 13998282. Bibcode: 1963PNAS...49..116W.
- "Establishing the Triplet Nature of the Genetic Code". Cell 128 (5): 815–818. 9 March 2007. doi:10.1016/j.cell.2007.02.029. PMID 17350564.
- "CodonExplorer: An Interactive Online Database for the Analysis of Codon Usage and Sequence Composition". Bioinformatics for DNA Sequence Analysis. Methods in Molecular Biology. 537. 28 February 2009. pp. 207–232. doi:10.1007/978-1-59745-251-9_10. ISBN 978-1-58829-910-9.
External links
Template:Featured list is only for Wikipedia:Featured lists.
 |