Software:SplitsTree
From HandWiki
| Developer(s) | Daniel Huson and David Bryant |
|---|---|
| Stable release | 4.16.0
/ 2020 |
| Operating system | Windows, Linux, Mac OS X |
| Type | Bioinformatics |
| License | Proprietary |
| Website | http://www.splitstree.org |
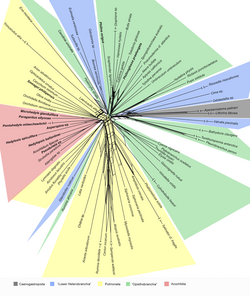
SplitsTree is a popular freeware program for inferring phylogenetic trees, phylogenetic networks, or, more generally, splits graphs, from various types of data such as a sequence alignment, a distance matrix or a set of trees.[1][2] SplitsTree implements published methods such as split decomposition,[3] neighbor-net, consensus networks,[4] super networks methods or methods for computing hybridization or simple recombination networks. It uses the NEXUS file format. The splits graph is defined using a special data block (SPLITS block).
See also
- Phylogenetic tree viewers
- Dendroscope
- MEGAN
References
- ↑ Dress, A.; K. T. Huber; V. Moulton (2001). "Metric spaces in pure and applied mathematics". Documenta Mathematica: 121–139. http://www.emis.ams.org/journals/DMJDMV/lsu/dress-huber-multon.pdf.
- ↑ Huson, D. H.; D. Bryant (2006). "Application of Phylogenetic Networks in Evolutionary Studies". Mol. Biol. Evol. 23 (2): 254–267. doi:10.1093/molbev/msj030. PMID 16221896.
- ↑ Bandelt, H. J.; Dress, A. W. (1992). "Split decomposition: a new and useful approach to phylogenetic analysis of distance data". Molecular Phylogenetics and Evolution 1 (3): 242–252. doi:10.1016/1055-7903(92)90021-8. ISSN 1055-7903. PMID 1342941.
- ↑ Holland, Barbara; Moulton, Vincent (2003). Benson, Gary; Page, Roderic D. M.. eds. "Consensus Networks: A Method for Visualising Incompatibilities in Collections of Trees" (in en). Algorithms in Bioinformatics. Lecture Notes in Computer Science (Springer Berlin Heidelberg) 2812: 165–176. doi:10.1007/978-3-540-39763-2_13. ISBN 9783540397632.
External links
- SplitsTree homepage (New Website for informations about SplitsTree)
- Alternative download page for the latest version (4.15) and manual (June 2019), hosted by the Department of Computer Science at the Eberhard Karls University Tübingen
- Algorithms in Bioinformatics, Daniel Huson's working group developing SplitsTree and other bioinformatics software
- List of phylogeny software, hosted at the University of Washington
- The Genealogical World of Phylogenetic Networks provides a wide range of examples for splits graphs, most of which were generated with SplitsTree
- Who is Who in Phylogenetic Networks lists software, researchers and literature dealing with phylogenetic networks
