Biology:Neighbor-net
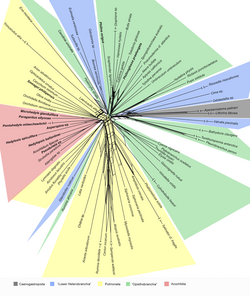
NeighborNet[1] is an algorithm for constructing phylogenetic networks which is loosely based on the neighbor joining algorithm. Like neighbor joining, the method takes a distance matrix as input, and works by agglomerating clusters. However, the NeighborNet algorithm can lead to collections of clusters which overlap and do not form a hierarchy, and are represented using a type of phylogenetic network called a splits graph. If the distance matrix satisfies the Kalmanson combinatorial conditions then Neighbor-net will return the corresponding circular ordering.[2][3] The method is implemented in the SplitsTree and R/Phangorn[4][5] packages.
Examples of the application of Neighbor-net can be found in virology,[6] horticulture,[7] dinosaur genetics,[8] comparative linguistics,[9] and archaeology.[10]
References
- ↑ "Neighbor-net: an agglomerative method for the construction of phylogenetic networks". Molecular Biology and Evolution 21 (2): 255–65. February 2004. doi:10.1093/molbev/msh018. PMID 14660700.
- ↑ "Consistency of the neighbor-net algorithm". Algorithms for Molecular Biology 2: 8. June 2007. doi:10.1186/1748-7188-2-8. PMID 17597551.
- ↑ "The neighbor-net algorithm.". Advances in Applied Mathematics 47 (2): 240–58. August 2011. doi:10.1016/j.aam.2010.09.002.
- ↑ "phangorn: phylogenetic analysis in R". Bioinformatics 27 (4): 592–3. February 2011. doi:10.1093/bioinformatics/btq706. PMID 21169378.
- ↑ "Intertwining phylogenetic trees and networks" (in en). Methods in Ecology and Evolution 8 (10): 1212–1220. 2017. doi:10.1111/2041-210X.12760.
- ↑ "A 10-year molecular survey of herpes simplex virus type 1 in Germany demonstrates a stable and high prevalence of genotypes A and B". Journal of Clinical Virology 44 (3): 235–7. March 2009. doi:10.1016/j.jcv.2008.12.016. PMID 19186100.
- ↑ "Independent wheat B and G genome origins in outcrossing Aegilops progenitor haplotypes". Molecular Biology and Evolution 24 (1): 217–27. January 2007. doi:10.1093/molbev/msl151. PMID 17053048.
- ↑ "Comment on "Protein sequences from mastodon and Tyrannosaurus rex revealed by mass spectrometry"". Science 319 (5859): 33; author reply 33. January 2008. doi:10.1126/science.1147046. PMID 18174420. Bibcode: 2008Sci...319...33B.
- ↑ Bowern, Claire (2010). "Historical linguistics in Australia: trees, networks and their implications". Philosophical Transactions of the Royal Society B: Biological Sciences 365 (1559): 3845–3854. doi:10.1098/rstb.2010.0013. ISSN 0962-8436. PMID 21041209.
- ↑ Pattern and process in cultural evolution.. University of California Press. 2009.
 |
