Biology:MicroDNA

MicroDNA is the most abundant subtype of Extrachromosomal Circular DNA (eccDNA) in humans, typically ranging from 200-400 base pairs in length and enriched in non-repetitive genomic sequences with a high density of exons.[2][3][4] Additionally, microDNA has been found to come from regions with CpG-islands which are commonly found within the 5' and 3' UTRs.[3][4][5] Being produced from regions of active transcription, it is hypothesized that microDNA may be formed as a by-product of transcriptional DNA damage repair.[5] MicroDNA is also thought to arise from other DNA repair pathways, mainly due to the parental sequences of microDNA having 2- to 15 bp direct repeats at the ends, resulting in replication slippage repair.[3] While only recently discovered, the role microDNA plays in and out of the cell is still not completely understood.[5] However, microDNA is currently thought to affect cellular homeostasis through transcription factor binding and have been used as a cancer biomarker.[5][6][7]
Discovery
MicroDNA was discovered through protocols similar to that of eccDNA extraction.[5] Specifically, eccDNA clones were generated through multiple displacement amplification and sequenced with Sanger sequencing, leading to microDNA's discovery.[5] Now with high-throughput sequencing being a more common practice, the complete genomic sequence of mammalian eccDNA has been obtained through the sequencing of the rolling amplification products of eccDNA.[5] Computational methods were then used to identify junctional sequences in the DNA.[4] The peaks found at lengths of 180 and 380 bp were discovered as microDNA and characterized by their CpG-islands and flanking 2- to 15 bp direct repeats.[4]
Since its discovery, microDNA has been identified in all tissue types and various samples, including mouse tissues and human cancer cell lines.[5][6] However, different species have unique genomic sites that specifically produce microDNA.[5] Because there are common genomic spots that produce microDNA in multiple cell and tissue types within a given species, there is evidence that they may not be produced solely as a DNA synthesis by-product.[5] However, studies have revealed separate clustering of microDNA extracted from cell-lines of different tissues, suggesting that formation may be linked to cell-lineage and unique transcriptional environments found in different cell types.[4][5]
Biogenesis
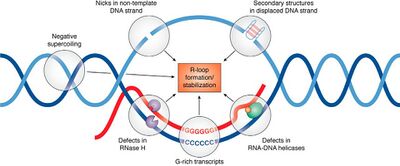
While the formation of microDNA is still uncertain, it has been linked to transcriptional activity and multiple DNA repair pathways.[3][5] As microDNA is produced from areas of high transcription activity/exon density, it could be formed from DNA repair during transcription.[5] Interestingly, triple-stranded DNA:RNA hybrids formed during transcription, termed R-loops, tend to form at CpG-islands within the 5' and 3' UTRs, similar to microDNA.[3][5] R-loops are correlated with DNA damage and genetic instability which is suggestive that microDNA may form from the single-stranded DNA (ssDNA) loop during the DNA damage response for R-loops.[3][5][6]
In DNA replication of short direct repeats (as found in the flanking regions of microDNA gene sources), it is possible for DNA loops to form, on the parent or product strand, through replication slippage.[3][5] To repair this, the mismatch repair (MMR) pathway can remove the loop and upon ligation of the repeating ends, single-strand microDNA can be produced.[3] The ss microDNA is then converted to double-stranded DNA; this process is still unknown.[5] It is important to note that if the loop is formed on the newly replicated strand, there is no consequential deletion in the genome while microdeletions can form from excisions in the template strand.[3][5] To understand the role MMR may have in microDNA biogenesis, analysis of microDNA abundance was performed in DT40 cells upon removal of MSH3, an essential protein in MMR.[3][5] The resulting microDNA from the DT40 MSH3-/- cell line had a higher enrichment of CpG-islands compared to the wild-type as well as an over 80% reduction of double-stranded microDNA.[3][5] Thus, it is hypothesized that the MMR pathway is essential for microDNA production from non-CpG islands in the genome while CpG enriched microDNA are formed by a different repair pathway.[3] File:DT40 microDNA.tif Again, because of the microhomology on the template genome, if there is a DNA break or a pause in replication (replication fork stalling), the newly synthesized DNA can circularize into ss microDNA.[3][5] This means when the template DNA is repaired after the creation of the microDNA, there is no deletion.[3]
MicroDNA created through the MMR pathway and replication fork stalling is a result of errors in DNA replication, however, there is evidence of microDNA being present in non-dividing cells as well.[5] This means that some microDNA is produced through repair pathways that also occur in quiescent cells, such as from 5' ends of LINE1 elements that are known to transpose.[3][5] To move around the genome, DNA transposons require transposase to remove the transposon from its original site and catalyze its insertion elsewhere in the genome.[3] Thus, the transposon is created by two double-stranded DNA breaks, also creating a microdeletion in the DNA.[3] This dsDNA fragment can be circularized through microhomology-mediated circularization, creating a ds microDNA.[3]
Implications
Transcription Factor Binding
Being 200-400 bp long, microDNA is too small to encode proteins, however, they may be important for molecular sponging.[4][5] Transcription factors often bind to promoter or regulatory sequences at the 5' end of DNA to initiate transcription.[5] These transcription factors can also bind to their respective recognition sites on microDNA because the microDNA often originates from the 5' UTRs of its parental gene, therefore, acting as a sponge for transcription factors.[4][5] This means microDNA can indirectly control gene expression and transcription homeostasis.[4][5]
Cancer Applications
In general, nucleic acid molecules that are found in the bloodstream, termed circulating or cell-free, are a relatively new disease biomarker being investigated, including for diagnosis and progression of cancer.[7] These molecules, such as cell-free DNA (cfDNA), are released into the blood upon cell death and in cases of cancer, can be identified based on the known mutations in oncogenes.[7]
Recent studies have extended the use of cell-free nucleic acids as cancer biomarkers to microDNA.[7] The cfmicroDNA was obtained from human and mouse serum and because of their similarities to cell-derived microDNA, as described above, it was concluded that cfmicroDNA is produced in the cell.[7] Similarly, when comparing lung tissue pre- and post-tumor removal, there was no found difference in circulating microDNA key characteristics other than an unexpected trend of longer circulating microDNA sequences in cancer patients pre-tumor removal.[7] The length of cfmicroDNA was found to be shorter post-surgery.[7]
Cell-free DNA is quickly cleared from the blood, making it a difficult cancer biomarker.[7] However, because circular DNA is not susceptible to DNA breakage by RNAse and exonuclease, it is more stable than linear DNA.[5][7] In combination with the observed lengthening of cfmicroDNA in cancer patient serum, this makes circulating microDNA a good cancer biomarker for both diagnosis and progression after treatment.[7]
See also
- Extrachromosomal DNA
- Extrachromosomal rDNA circle
- Selfish genetic elements
- Double minute
References
- ↑ "File:CpG vs C-G bp.svg - Wikipedia" (in en). 31 January 2016. https://commons.wikimedia.org/wiki/File:CpG_vs_C-G_bp.svg.
- ↑ "Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues". Science 336 (6077): 82–86. April 2012. doi:10.1126/science.1213307. PMID 22403181. Bibcode: 2012Sci...336...82S.
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 3.11 3.12 3.13 3.14 3.15 3.16 3.17 "Production of Extrachromosomal MicroDNAs Is Linked to Mismatch Repair Pathways and Transcriptional Activity". Cell Reports 11 (11): 1749–1759. June 2015. doi:10.1016/j.celrep.2015.05.020. PMID 26051933.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 4.7 "Discoveries of Extrachromosomal Circles of DNA in Normal and Tumor Cells". Trends in Genetics 34 (4): 270–278. April 2018. doi:10.1016/j.tig.2017.12.010. PMID 29329720.
- ↑ 5.00 5.01 5.02 5.03 5.04 5.05 5.06 5.07 5.08 5.09 5.10 5.11 5.12 5.13 5.14 5.15 5.16 5.17 5.18 5.19 5.20 5.21 5.22 5.23 5.24 5.25 5.26 5.27 Reon, Brian J.; Dutta, Anindya (2016-04-01). "Biological Processes Discovered by High-Throughput Sequencing" (in en). The American Journal of Pathology 186 (4): 722–732. doi:10.1016/j.ajpath.2015.10.033. ISSN 0002-9440. PMID 26828742.
- ↑ 6.0 6.1 6.2 "Normal and Cancerous Tissues Release Extrachromosomal Circular DNA (eccDNA) into the Circulation". Molecular Cancer Research 15 (9): 1197–1205. September 2017. doi:10.1158/1541-7786.MCR-17-0095. PMID 28550083. PMC 5581709. https://mcr.aacrjournals.org/content/15/9/1197.
- ↑ 7.0 7.1 7.2 7.3 7.4 7.5 7.6 7.7 7.8 7.9 Kumar, Pankaj; Dillon, Laura W.; Shibata, Yoshiyuki; Jazaeri, Amir A.; Jones, David R.; Dutta, Anindya (2017-09-01). "Normal and Cancerous Tissues Release Extrachromosomal Circular DNA (eccDNA) into the Circulation" (in en). Molecular Cancer Research 15 (9): 1197–1205. doi:10.1158/1541-7786.MCR-17-0095. ISSN 1541-7786. PMID 28550083. PMC 5581709. https://mcr.aacrjournals.org/content/15/9/1197.
- ↑ "File:R-loop promoting factors.jpg - Wikipedia" (in en). 24 January 2021. https://commons.wikimedia.org/wiki/File:R-loop_promoting_factors.jpg.
- ↑ "File:DT40 microDNA.tif - Wikipedia" (in en). 20 March 2015. https://commons.wikimedia.org/wiki/File:DT40_microDNA.tif.
 |

