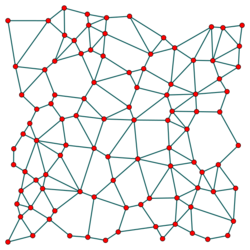
Gabriel graph
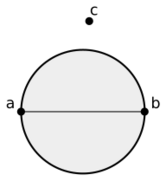
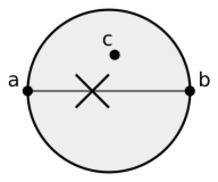
In mathematics and computational geometry, the Gabriel graph of a set [math]\displaystyle{ S }[/math] of points in the Euclidean plane expresses one notion of proximity or nearness of those points. Formally, it is the graph [math]\displaystyle{ G }[/math] with vertex set [math]\displaystyle{ S }[/math] in which any two distinct points [math]\displaystyle{ p \in S }[/math] and [math]\displaystyle{ q \in S }[/math] are adjacent precisely when the closed disc having [math]\displaystyle{ pq }[/math] as a diameter contains no other points. Another way of expressing the same adjacency criterion is that [math]\displaystyle{ p }[/math] and [math]\displaystyle{ q }[/math] should be the two closest given points to their midpoint, with no other given point being as close. Gabriel graphs naturally generalize to higher dimensions, with the empty disks replaced by empty closed balls. Gabriel graphs are named after K. Ruben Gabriel, who introduced them in a paper with Robert R. Sokal in 1969.[1]
Percolation
For Gabriel graphs of infinite random point sets, the finite site percolation threshold gives the fraction of points needed to support connectivity: if a random subset of fewer vertices than the threshold is given, the remaining graph will almost surely have only finite connected components, while if the size of the random subset is more than the threshold, then the remaining graph will almost surely have an infinite component (as well as finite components). This threshold was proved to exist by (Bertin Billiot),[2] and more precise values of both site and bond thresholds have been given by Norrenbrock.[3]
Related geometric graphs
The Gabriel graph is a subgraph of the Delaunay triangulation. It can be found in linear time if the Delaunay triangulation is given.[4]
The Gabriel graph contains, as subgraphs, the Euclidean minimum spanning tree, the relative neighborhood graph, and the nearest neighbor graph.
It is an instance of a beta-skeleton. Like beta-skeletons, and unlike Delaunay triangulations, it is not a geometric spanner: for some point sets, distances within the Gabriel graph can be much larger than the Euclidean distances between points.[5]
References
- ↑ "A new statistical approach to geographic variation analysis", Systematic Biology 18 (3): 259–278, 1969, doi:10.2307/2412323
- ↑ Bertin, Etienne; Billiot, Jean-Michel; Drouilhet, Rémy (2002), "Continuum percolation in the Gabriel graph", Advances in Applied Probability 34 (4): 689–701, doi:10.1239/aap/1037990948
- ↑ Norrenbrock, Christoph (May 2016), "Percolation threshold on planar Euclidean Gabriel graphs", European Physical Journal B 89 (5): 111, doi:10.1140/epjb/e2016-60728-0, Bibcode: 2016EPJB...89..111N
- ↑ "Properties of Gabriel graphs relevant to geographic variation research and clustering of points in the plane", Geographical Analysis 12 (3): 205–222, 1980, doi:10.1111/j.1538-4632.1980.tb00031.x
- ↑ "On the spanning ratio of Gabriel graphs and β-skeletons", SIAM Journal on Discrete Mathematics 20 (2): 412–427, 2006, doi:10.1137/S0895480197318088
 |