Biology:KRBA1
 Generic protein structure example |
KRBA1 is a protein that in humans is encoded by the KRBA1 gene. It is located on the plus strand of chromosome 7 from 149,411,872 to 149,431,664.[1] It is also commonly known under two other aliases: KIAA1862 and KRAB A Domain Containing 1 gene and encodes the KRBA1 protein in humans.[2] The KRBA family of genes is understood to encode different transcriptional repressor proteins[3]
Gene
The KRBA1 gene is located on chromosome 7 at 7q36 in humans. It is 3786 base pairs in length with 17 exons total, however the coding sequence is only 3196 base pairs long resulting in a protein with 1064 amino acids.[4]
Protein
The KRBA1 protein has a predicted isoelectric point of 7.8pI and a predicted molecular weight of 112 kD which makes it a relatively large and neutral protein.[5] This protein is primarily made up of four amino acids. The four major ones are: Proline at 15.7%, Glycine at 11.2%, Serine at 10.9%, and Leucine at 10.2%.[5] Proline is present in well above average proportions. However, it was the only one as such and the amino acids M, N, Y, I, and F are all present in just below average proportions.
Isoforms
There are two isoforms for this protein known simply as isoform 1 and 2. Isoform 1 is the smaller one as it has a shorter N-terminus. Additionally, its 5'-UTR is different and its start codon is farther downstream.[4]
Domains and motifs
The domain of function is the KRBA box which is predicted to start at base pair 7 and end at base pair 47. For motifs there was a leucine zipper and two nuclear localization signals (NLS) were found.[6] Additionally, two fairly large proline rich regions are present towards the C-terminus of the protein.[7]
The leucine zipper is present at base pair 837: LHSLGAALAEKLDRLATALAGL
One 4 pattern and one 7 pattern NLS were found
pat4: KRPR @ 763bp
pat7: PSRRKSH @ 217bp
Secondary and tertiary structure
Using the GOR4 program, it can be inferred that KRBA1 has very little specific secondary structure composed of mostly random coil regions. Random coil regions constitute 75.00% of the protein, while alpha helices constitute 16.26% and extended strands 8.74%.[8] The alpha helices are broken up into many sections throughout the protein.
DNA level regulation
Promoter
The promoter region was chosen using ElDorado at Genomatix, which assessed the KRBA1 gene locus for possible promoter regions. Out of nine possible promoter sets, Promoter Set 2 (GXP_660502) was chosen, as its transcripts had much higher numbers of CAGE tags than any other set. Coding transcript GXT_27212195 was used, as it had the highest number of exons (17/17 possible), and matched with the NM_001290187 accession number on NCBI for KRBA1 mRNA.[9]
Transcription factor binding sites
| Matrix Family | Detailed Family Information | Matrix | Detailed Matrix Information | Starting bp | Ending bp | Anchor bp | Strand | Similarity | Sequence |
|---|---|---|---|---|---|---|---|---|---|
| V$ZTRE | Zinc transcriptional regulatory element | V$ZTRE.03 | 5' half site of ZTRE motif | 1141 | 1157 | 1149 | (+) | 0.993 | ccCTCCccctcggccca |
| V$ZTRE | Zinc transcriptional regulatory element | V$ZTRE.03 | 5' half site of ZTRE motif | 898 | 914 | 906 | (-) | 0.989 | ccCTCCcctcgtccccg |
| V$ZTRE | Zinc transcriptional regulatory element | V$ZTRE.04 | 3' half site of ZTRE motif | 1133 | 1149 | 1141 | (-) | 0.984 | gggGGAGgggcgggggt |
| V$ZTRE | Zinc transcriptional regulatory element | V$ZTRE.04 | 3' half site of ZTRE motif | 906 | 922 | 914 | (+) | 0.983 | aggGGAGggggcgggga |
| V$PLAG | Pleomorphic adenoma gene | V$PLAG1.01 | Pleomorphic adenoma gene (PLAG) 1, a developmentally regulated C2H2 zinc finger protein | 66 | 88 | 77 | (+) | 0.951 | cgGAGGccgggacgaggggttgt |
| V$PLAG | Pleomorphic adenoma gene | V$PLAG1.02 | Pleomorphic adenoma gene 1 | 307 | 329 | 318 | (-) | 1 | aaGGGGgcctgcggggctaagcc |
| V$PLAG | Pleomorphic adenoma gene | V$PLAG1.02 | Pleomorphic adenoma gene 1 | 459 | 481 | 470 | (+) | 1 | caGGGGgaggcagaggagagtgg |
| V$PLAG | Pleomorphic adenoma gene | V$PLAG1.02 | Pleomorphic adenoma gene 1 | 30 | 52 | 41 | (+) | 1 | gaGGGGgctttgcccgagtgggc |
| V$SMAD | Vertebrate SMAD family of transcription factors | V$SMAD3.01 | Smad3 transcription factor involved in TGF-beta signaling | 199 | 209 | 204 | (+) | 0.994 | gctGTCTgggc |
| V$SMAD | Vertebrate SMAD family of transcription factors | V$SMAD3.01 | Smad3 transcription factor involved in TGF-beta signaling | 213 | 223 | 218 | (+) | 0.996 | aagGTCTggac |
| V$SMAD | Vertebrate SMAD family of transcription factors | V$SMAD3.01 | Smad3 transcription factor involved in TGF-beta signaling | 428 | 438 | 433 | (-) | 0.994 | ctgGTCTgggc |
| V$SMAD | Vertebrate SMAD family of transcription factors | V$SMAD3.02 | Smad3 transcription factor involved in TGF-beta signaling factor PU.1 | 363 | 373 | 368 | (-) | 0.993 | cctGTCTggag |
| V$AHRR | AHR-arnt heterodimers and AHR-related factors | V$AHRARNT.03 | DRE (dioxin response elements), XRE (xenobiotic response elements) bound by AHR/ARNT heterodimers | 41 | 65 | 53 | (+) | 0.971 | gcccgagtggGCGTgcgcctttcct |
| V$AHRR | AHR-arnt heterodimers and AHR-related factors | V$AHRARNT.03 | DRE (dioxin response elements), XRE (xenobiotic response elements) bound by AHR/ARNT heterodimers | 246 | 270 | 258 | (+) | 0.952 | agtctcctctGCGTgggaccacagc |
| V$AHRR | AHR-arnt heterodimers and AHR-related factors | V$AHRARNT.01 | Aryl hydrocarbon receptor / Arnt heterodimers | 556 | 580 | 568 | (-) | 0.97 | gtcacattttgCGTGcctgtttgct |
| V$AHRR | AHR-arnt heterodimers and AHR-related factors | V$AHRARNT.03 | DRE (dioxin response elements), XRE (xenobiotic response elements) bound by AHR/ARNT heterodimers | 658 | 682 | 670 | (-) | 0.956 | gtccggccggGCGTgggtgggacag |
Expression pattern
In humans, this gene is ubiquitously expressed at a fairly low level, approximately 0.6 times the expression of the average gene.[4] Keeping that in mind, this gene is expressed the most in the heart. There is some expression in other areas such as the reproductive organs and the brain, but it is much less. When it comes to the common mouse Mus musculus, the expression of the KRBA1 gene occurs in different areas of the body compared to humans. In mice, the highest expression isn't in the heart but in the reproductive organs along with the adrenal gland and thymus.[4]
Protein level regulation
Localization
This protein is localized in the nucleus.[1] The gene is predicted to contain two nuclear localization signals (NLS) both a 4 residue pattern and 7 residue pattern signal.[6]
Post translational modifications
There weren't many different types of post translational modifications that were found. However, the few that were found occur throughout the protein at high levels. The few that were found were: O-GlcNAc,[10] O-Glycosylation,[11] and net phosphorylation,[12] and glycation.[13] All four of them have a multitude of sites throughout the entire protein.
Homology and evolution
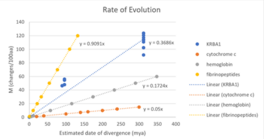
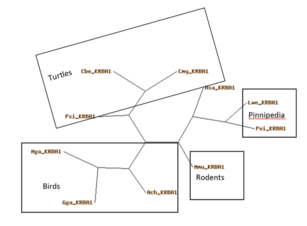
KRBA1 orthologs have a relatively high mutation rate as seen in the graph below comparing KRBA1 with fibrinopeptides, hemoglobin, and cytochrome C. The orthologs in the table below are sorted by sequence identity and similarity.[4]
| Genus and Species | Common name | Taxonomic Group | Date of divergence
(MYA) |
Accession number | Sequence length (aa) | Sequence identity % | Sequence similarity |
| Homo sapiens | Humans | primates | n/a | NP_001277116.1 | 1064 | 100% | 100% |
| Equus asinus | Asinus | Equidae | 96 | XP_014686007.1 | 1179 | 63% | 69% |
| Leptonychotes weddellii | Weddell seal | Pinnipedia | 96 | XP_030881320.1 | 1142 | 63% | 69% |
| Phoca vitulina | Harbor seal | Pinnipedia | 96 | XP_032257255.1 | 1184 | 63% | 69% |
| Nannospalax galili | Greater mole rat | Rodentia | 90 | XP_008832018.1 | 1103 | 62% | 69% |
| Felis catus | Cat | Felidae | 96 | XP_011278956.2 | 1159 | 62% | 67% |
| Mus musculus | Mouse | Rodentia | 90 | NP_001334081.1 | 1078 | 58% | 67% |
| Mus Pahari | Garidner's shrewmouse | Rodentia | 90 | XP_029390880.1 | 1086 | 57% | 66% |
| Gallus gallus | Chicken | Aves | 312 | XP_025003155.1 | 555 | 40% | 52% |
| Pogona vitticeps | Central bearded dragon | Reptilia | 312 | XP_020655832.1 | 1697 | 36% | 51% |
Function and biochemistry
The domain of function on this gene is known as the KRAB (Kruppel-associated box) domain - A box and it spans from amino acid 7 to 47. It is a transcription repression domain within the zinc finger proteins subgroup of the KRAB-ZFPs family.[3] The KRAB domain interacts with and recruits other proteins and corepressors and then represses transcription. The mechanism appears to be the following: the KRBA proteins via their KRAB domain recruit the repressor KAP1 (KRAB-associated protein-1, also known as transcription intermediary factor 1 beta, KRAB-A interacting protein). The KAP1/ KRAB complex then recruits the heterochromatin protein 1 (HP1), and other chromatin modulating proteins, leading to transcriptional repression through heterochromatin formation.[3]
Interacting proteins
Using the STRING database, two interacting proteins were found to have high confidence interaction scores. High interaction scores are any scores above 0.7. The two proteins are Transcription intermediary factor 1-beta (TRIM28) with a score of 0.903 and Leucine-rich repeat-containing protein 61 (LRRC61) with a score 0.803.[15]
Clinical significance
A research done by Zaibo Li et al. displayed the KRAB-A-Domain in action. The von-Hippel Lindau tumor suppressor (pVHL) is a part of protein complex known as E3 ubiquitin ligase and targets hypoxia-inducible-factor 1a (HIF-1a) for breakdown. It is reported that there is a new protein that was discovered that interacts with the pVHL protein and helps it with its job.[16] pVHL works by bringing in histone deacetylases, and it is helped by a certain domain on this new protein. It is shown in a study by Li et al. that this domain is the KRAB-A-domain and it mediates pVHL's transcriptional repression of HIF-1a.[16] Additionally, the research shows that the KRAB-A-domain is shown to repress transcription on top of its ability to recruit histone deacetylases.[16]
References
- ↑ 1.0 1.1 "AceView: Gene:KRBA1, a comprehensive annotation of human, mouse and worm genes with mRNAs or ESTsAceView.". https://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/av.cgi?db=human&q=KRBA1.
- ↑ "KRBA1 Gene - GeneCards | KRBA1 Protein | KRBA1 Antibody". https://www.genecards.org/cgi-bin/carddisp.pl?gene=KRBA1.
- ↑ 3.0 3.1 3.2 "CDD Conserved Protein Domain Family: KRAB_A-box". https://www.ncbi.nlm.nih.gov/Structure/cdd/cl02581.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 "KRBA1 KRAB-A domain containing 1 [Homo sapiens (human) - Gene - NCBI"]. https://www.ncbi.nlm.nih.gov/gene?cmd=retrieve&list_uids=84626.
- ↑ 5.0 5.1 "ExPASy - Compute pI/Mw tool". https://web.expasy.org/compute_pi/.
- ↑ 6.0 6.1 "PSORT II Prediction". https://psort.hgc.jp/form2.html.
- ↑ "ELM - Search the ELM resource". http://elm.eu.org/.
- ↑ "NPS@ : GOR4 secondary structure prediction". https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_gor4.html.
- ↑ 9.0 9.1 "Genomatix - NGS Data Analysis & Personalized Medicine". https://www.genomatix.de/?s=14e3197a7642d3f12be8bd8a30214856.
- ↑ "YinOYang 1.2 Server". http://www.cbs.dtu.dk/services/YinOYang/.
- ↑ "NetOGlyc 4.0 Server". http://www.cbs.dtu.dk/services/NetOGlyc/.
- ↑ "NetPhos 3.1 Server". http://www.cbs.dtu.dk/services/NetPhos/.
- ↑ "NetGlycate 1.0 Server". http://www.cbs.dtu.dk/services/NetGlycate/.
- ↑ "TimeTree :: The Timescale of Life". http://www.timetree.org/.
- ↑ "KRBA1 protein (human) - STRING interaction network". https://version11.string-db.org/cgi/network.pl?taskId=xKw7HUvfVj8S.
- ↑ 16.0 16.1 16.2 "The VHL protein recruits a novel KRAB-A domain protein to repress HIF-1alpha transcriptional activity". The EMBO Journal 22 (8): 1857–67. April 2003. doi:10.1093/emboj/cdg173. PMID 12682018.
 |