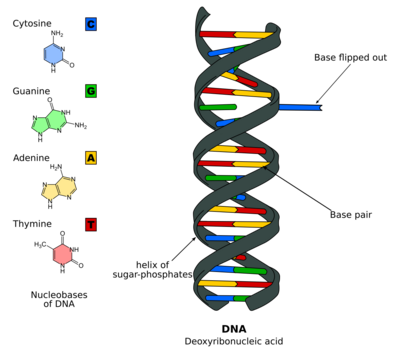
Biology:DNA base flipping
DNA base flipping, or nucleotide flipping, is a mechanism in which a single nucleotide base, or nucleobase, is rotated outside the nucleic acid double helix.[1] This occurs when a nucleic acid-processing enzyme needs access to the base to perform work on it, such as its excision for replacement with another base during DNA repair. It was first observed in 1994 using X-ray crystallography in a methyltransferase enzyme catalyzing methylation of a cytosine base in DNA. Since then, it has been shown to be used by different enzymes in many biological processes such as DNA methylation, various DNA repair mechanisms, and DNA replication. It can also occur in RNA double helices [2] or in the DNA:RNA intermediates formed during RNA transcription.
DNA base flipping occurs by breaking the hydrogen bonds between the bases and unstacking the base from its neighbors. This could occur through an active process, where an enzyme binds to the DNA and then facilitates rotation of the base, or a passive process, where the base rotates out spontaneously, and this state is recognized and bound by an enzyme. It can be detected using X-ray crystallography, NMR spectroscopy, fluorescence spectroscopy, or hybridization probes.
Discovery
Base flipping was first observed in 1994 when researchers Klimasauskas, Kumar, Roberts, and Cheng used X-ray crystallography to view an intermediate step in the chemical reaction of a methyltransferase bound to DNA.[3] The methyltransferase they used was the C5-cytosine methyltransferase from Haemophilus haemolyticus (M. HhaI). This enzyme recognizes a specific sequence of the DNA (5'-GCGC-3') and methylates the first cytosine base of the sequence at its C5 location.[3] Upon crystallization of the M. HhaI-DNA complex, they saw the target cytosine base was rotated completely out of the double helix and was positioned in the active site of the M. HhaI. It was held in place by numerous interactions between the M. HhaI and DNA.[3]
The authors theorized that base flipping was a mechanism used by many other enzymes, such as helicases, recombination enzymes, RNA polymerases, DNA polymerases, and Type II topoisomerases.[3] Much research has been done in the years subsequent to this discovery and it has been found that base flipping is a mechanism used in many of the biological processes the authors suggest.[4][5][6]
Mechanism
DNA nucleotides are held together with hydrogen bonds, which are relatively weak and can be easily broken. Base flipping occurs on a millisecond timescale[7] by breaking the hydrogen bonds between bases and unstacking the base from its neighbors.[8] The base is rotated out of the double helix by 180 degrees.,[9] typically via the major groove,[10] and into the active site of an enzyme. This opening leads to small conformational changes in the DNA backbone[11] which are quickly stabilized by the increased enzyme-DNA interactions.[12] Studies looking at the free-energy profiles of base flipping have shown that the free-energy barrier to flipping can be lowered by 17 kcal/mol for M.HhaI in the closed conformation.[10]
There are two mechanisms of DNA base flipping: active and passive.[13] In the active mechanism, an enzyme binds to the DNA and then actively rotates the base, while in the passive mechanism a damaged base rotates out spontaneously first, then is recognized and bound by the enzyme.[8] Research has demonstrated both mechanisms: uracil-DNA glycosylase follows the passive mechanism[8] and Tn10 transposase follows the active mechanism.[14]
Furthermore, studies have shown that DNA base flipping is used by many different enzymes in a variety biological processes such as DNA methylation, various DNA repair mechanisms, RNA transcription and DNA replication.[4][5][6]
Biological processes
DNA modification and repair
DNA can have mutations that cause a base in the DNA strand to be damaged. To ensure genetic integrity of the DNA, enzymes need to repair any damage. There are many types of DNA repair. Base excision repair utilizes base flipping to flip the damaged base out of the double helix[5] and into the specificity pocket of a glycosylase which hydrolyzes the glycosidic bond and removes the base.[15] DNA glycosylases interact with DNA, flipping bases to determine a mismatch. An example of base excision repair occurs when a cytosine base is deaminated and becomes a uracil base. This causes a U:G mispair which is detected by Uracil DNA glycosylase. The uracil base is flipped out into the glycosylase active pocket where it is removed from the DNA strand.[16] Base flipping is used to repair mutations such as 8-Oxoguanine (oxoG)[17] and thymine dimers created by UV radiation.[15][18]
Replication, transcription and recombination
DNA replication and RNA transcription both make use of base flipping.[5] DNA polymerase is an enzyme that carries out replication. It can be thought of as a hand that grips the DNA single strand template.[15] As the template passes across the palm region of the polymerase, the template bases are flipped out of the helix and away from the dNTP binding site.[19] During transcription, RNA polymerase catalyzes RNA synthesis. During the initiation phase, two bases in the -10 element flip out from the helix and into two pockets in RNA polymerase. These new interactions stabilize the -10 element and promote the DNA strands to separate or melt.[15][20]
Base flipping occurs during latter stages of recombination.[21] RecA is a protein that promotes strand invasion[15] during homologous recombination. Base flipping has been proposed as the mechanism by which RecA can enable a single strand to recognize homology in duplex DNA.[22] Other studies indicate that it is also involved in V(D)J Recombination.[23]
DNA methylation
DNA methylation is the process in which a methyl group is added to either a cytosine or adenine.[24] This process causes the activation or inactivation of gene expression, thereby resulting in gene regulation in eukaryotic cells. DNA methylation process is also known to be involved in certain types of cancer formation.[25][26][27] In order for this chemical modification to occur, it is necessary that the target base flips out of the DNA double helix to allow the methyltransferases to catalyze the reaction.[5]
Target recognition by restriction endonucleases
Restriction endonucleases, also known as restriction enzymes are enzymes that cleave the sugar-phosphate backbone of the DNA at specific nucleotides sequences that are usually four to six nucleotides long.[28] Studies performed by Horton and colleagues have shown that the mechanism by which these enzymes cleave the DNA involves base flipping as well as bending the DNA and the expansion of the minor groove.[29] In 2006, Horton and colleagues, x-ray crystallography evidence was presented showing that the restriction endonuclease HinP1I utilizes base flipping in order to recognize its target sequence. This enzyme is known to cleave the DNA at the palindromic tetranucleotide sequence G↓CGC.
Experimental approaches for detection
X-ray crystallography
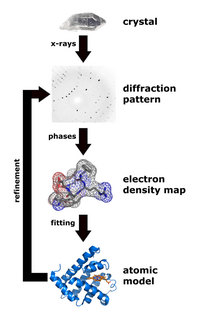
X-ray crystallography is a technique that measures the angles and intensities of crystalline atoms in order to determine the atomic and molecular structure of the crystal of interest. Crystallographers are then able to produce and three-dimensional picture where the positions of the atoms, chemical bonds as well as other important characteristics can be determined.[30] Klimasaukas and colleagues used this technique to observe the first base flipping phenomenon, in which their experimental procedure involved several steps:[3]
- Purification
- Crystallization
- Data Collection
- Structure determination and refinement
During purification, Haemophilus haemolyticus methyltransferase was overexpressed and purified using a high salt back-extraction step to selectively solubilize M.HhaI, followed by fast protein liquid chromatography (FPLC) as done previously by Kumar and colleagues.[31] Authors utilized a Mono-Q anion exchange column to remove the small quantity of proteinaceous materials and unwanted DNA prior to the crystallization step. Once M.HhaI was successfully purified, the sample was then grown using a method that mixes the solution containing the complex at a temperature of 16 °C and the hanging-drop vapor diffusion technique to obtain the crystals. Authors were then able to collect the x-ray data according to a technique used by Cheng and colleagues in 1993.[32] This technique involved the measurement of the diffraction intensities on a FAST detector, where the exposure times for 0.1° rotation were 5 or 10 seconds. For the structure determination and refinement, Klimasaukas and colleagues used the molecular replacement of the refined apo structure described by Cheng and colleagues in 1993[32] where the search models X-PLOR, MERLOT, and TRNSUM were used to solve the rotation and translation functions.[33][34] This part of the study involves the use of a variety of software and computer algorithms to solve the structures and characteristics of the crystal of interest.
NMR spectroscopy
NMR spectroscopy is a technique that has been used over the years to study important dynamic aspects of base flipping. This technique allows researchers to determine the physical and chemical properties of atoms and other molecules by utilizing the magnetic properties of atomic nuclei.[35] In addition, NMR can provide a variety of information including structure, reaction states, chemical environment of the molecules, and dynamics.[36][37] During the DNA base flipping discovery experiment, researchers utilized NMR spectroscopy to investigate the enzyme-induced base flipping of HhaI methyltransferase. In order to accomplish this experiment, two 5-fluorocytosine residues were incorporated into the target and the reference position with the DNA substrate so the 19F chemical shift analysis could be performed. Once the 19F chemical shift analysis was evaluated, it was then concluded that the DNA complexes existed with multiple forms of the target 5-fluorocytosine along the base flipping pathway.[38]
Fluorescence spectroscopy
Fluorescence spectroscopy is a technique that is used to assay a sample using a fluorescent probe. DNA nucleotides themselves are not good candidates for this technique because they do not readily re-emit light upon light excitation.[39] A fluorescent marker is needed to detect base flipping. 2-Aminopurine is a base that is structurally similar to adenine, but is very fluorescent when flipped out from the DNA duplex.[40] It is commonly used to detect base flipping and has an excitation at 305‑320 nm and emission at 370 nm so that it well separated from the excitations of proteins and DNA. Other fluorescent probes used to study DNA base flipping are 6MAP (4‑amino‑6‑methyl‑7(8H)‑pteridone)[41] and Pyrrolo‑C (3-[β-D-2-ribofuranosyl]-6-methylpyrrolo[2,3-d]pyrimidin-2(3H)-one).[42][43] Time-resolved fluorescence spectroscopy is also employed to provide a more detailed picture of the extent of base flipping as well as the conformational dynamics occurring during base flipping.[44]
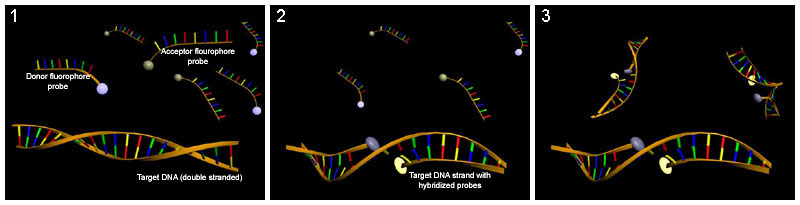
Hybridization probing
Hybridization probes can be used to detect base flipping. This technique uses a molecule that has a complementary sequence to the sequence you would like to detect such that it binds to a single-strand of the DNA or RNA. Several hybridization probes have been used to detect base flipping. Potassium permanganate is used to detect thymine residues that have been flipped out by cytosine-C5 and adenine-N6 methyltransferases.[45] Chloroacetaldehyde is used to detect cytosine residues flipped out by the HhaI DNA cytosine-5 methyltransferase (M. HhaI).[46]
See also
- DNA repair
- Base excision repair
- DNA replication
- RNA transcription
- DNA methylation
- DNA methyltransferase
- Genetic recombination
- Homologous recombination
- DNA
- Epigenetics
- Epigenomics
References
- ↑ Roberts, RJ; Cheng, X (1998). "Base flipping". Annual Review of Biochemistry 67 (1): 181–198. doi:10.1146/annurev.biochem.67.1.181. PMID 9759487.
- ↑ Reiter, NJ; Blad, H; Abildgaard, F; Butcher, SE (2004). "Dynamics in the U6 RNA intramolecular stem-loop: a base flipping conformational change". Biochemistry 43 (43): 13739–47. doi:10.1021/bi048815y. PMID 15504036.
- ↑ 3.0 3.1 3.2 3.3 3.4 Klimasauskas, Saulius; Kumar, Sanjay; Roberts, Richard J.; Cheng, Xiaodong (January 1994). "Hhal methyltransferase flips its target base out of the DNA helix". Cell 76 (2): 357–369. doi:10.1016/0092-8674(94)90342-5. PMID 8293469.
- ↑ 4.0 4.1 Brown, Tom. "Nucleic Acids Book". http://www.atdbio.com/content/54/Base-flipping#Further-reading.
- ↑ 5.0 5.1 5.2 5.3 5.4 Huang, Niu; Nilesh K. Banavali; Alexander D. MacKerell (January 7, 2003). "Protein-facilitated base flipping in DNA by cytosine-5-methyltransferase". PNAS 100 (1): 68–73. doi:10.1073/pnas.0135427100. PMID 12506195. Bibcode: 2003PNAS..100...68H.
- ↑ 6.0 6.1 Grubmüller, Helmut. "DNA Base Flipping". http://www.mpibpc.mpg.de/259673/10-DNA-Base-Flipping.
- ↑ Bouvier, Benjamin; Grubmüller, Helmut (August 2007). "A Molecular Dynamics Study of Slow Base Flipping in DNA using Conformational Flooding". Biophysical Journal 93 (3): 770–786. doi:10.1529/biophysj.106.091751. PMID 17496048. PMC 1913169. Bibcode: 2007BpJ....93..770B. http://www.mpibpc.mpg.de/276493/paper_moleculare_dynamics_study.pdf. Retrieved 2014-03-15.
- ↑ 8.0 8.1 8.2 Lariviere, L. (23 June 2004). "Structural Evidence of a Passive Base-flipping Mechanism for Glucosyltransferase". Journal of Biological Chemistry 279 (33): 34715–34720. doi:10.1074/jbc.M404394200. PMID 15178685.
- ↑ Grosjean, [edited by] Henri (2009). DNA and RNA modification enzymes : structure, mechanism, function and evolution. Austin, Tex.: Landes Bioscience. ISBN 978-1-58706-329-9. https://www.landesbioscience.com/curie/chapter/4163/. Retrieved 2014-03-10.
- ↑ 10.0 10.1 Huang, N.; Banavali, N. K.; MacKerell, A. D. (27 December 2002). "Protein-facilitated base flipping in DNA by cytosine-5-methyltransferase". Proceedings of the National Academy of Sciences 100 (1): 68–73. doi:10.1073/pnas.0135427100. PMID 12506195.
- ↑ Giudice, E. (1 March 2003). "Base pair opening within B-DNA: free energy pathways for GC and AT pairs from umbrella sampling simulations". Nucleic Acids Research 31 (5): 1434–1443. doi:10.1093/nar/gkg239. PMID 12595551.
- ↑ Huang, N.; Banavali, N. K.; MacKerell, A. D. (27 December 2002). "Protein-facilitated base flipping in DNA by cytosine-5-methyltransferase". Proceedings of the National Academy of Sciences 100 (1): 68–73. doi:10.1073/pnas.0135427100. PMID 12506195.
- ↑ O'Neil, Lauren. Base Flipping in DNA: Detection, Structures and Energetics, A Dissertation. ISBN 9780549590743. https://books.google.com/books?id=Zm26mDpW3TIC. Retrieved 15 March 2014.[yes|permanent dead link|dead link}}]
- ↑ Bischerour, Julien; Chalmers, Ronald; Bielinsky, Anja-Katrin (10 July 2009). "Base Flipping in Tn10 Transposition: An Active Flip and Capture Mechanism". PLOS ONE 4 (7): e6201. doi:10.1371/journal.pone.0006201. PMID 19593448. Bibcode: 2009PLoSO...4.6201B.
- ↑ 15.0 15.1 15.2 15.3 15.4 University, James D. Watson, Cold Spring Harbor Laboratory, Tania A. Baker, Massachusetts Institute of Technology, Alexander Gann, Cold Spring Harbor Laboratory, Michael Levine, University of California, Berkeley, Richard Losik, Harvard (2014). Molecular biology of the gene (Seventh ed.). Boston: Pearson/CSH Press. ISBN 978-0-321-76243-6.
- ↑ Krokan, Hans E; Drabløs, Finn; Slupphaug, Geir (16 December 2002). "Uracil in DNA – occurrence, consequences and repair". Oncogene 21 (58): 8935–8948. doi:10.1038/sj.onc.1205996. PMID 12483510.
- ↑ Banerjee, Anirban; Yang, Wei; Karplus, Martin; Verdine, Gregory L. (31 March 2005). "Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA". Nature 434 (7033): 612–618. doi:10.1038/nature03458. PMID 15800616. Bibcode: 2005Natur.434..612B.
- ↑ Fuxreiter, Monika; Luo, Ning; Jedlovszky, Pál; Simon, István; Osman, Roman (November 2002). "Role of Base Flipping in Specific Recognition of Damaged DNA by Repair Enzymes". Journal of Molecular Biology 323 (5): 823–834. doi:10.1016/S0022-2836(02)00999-3. PMID 12417196.
- ↑ Patel, Premal H.; Suzuki, Motoshi; Adman, Elinor; Shinkai, Akeo; Loeb, Lawrence A. (May 2001). "Prokaryotic DNA polymerase I: evolution, structure, and "base flipping" mechanism for nucleotide selection". Journal of Molecular Biology 308 (5): 823–837. doi:10.1006/jmbi.2001.4619. PMID 11352575.
- ↑ Lim, H. M.; Lee, H. J.; Roy, S.; Adhya, S. (4 December 2001). "A "master" in base unpairing during isomerization of a promoter upon RNA polymerase binding". Proceedings of the National Academy of Sciences 98 (26): 14849–14852. doi:10.1073/pnas.261517398. PMID 11734629. Bibcode: 2001PNAS...9814849L.
- ↑ Voloshin, Oleg N.; Camerini-Otero, R.Daniel (September 2004). "Synaptic Complex Revisited". Molecular Cell 15 (6): 846–847. doi:10.1016/j.molcel.2004.09.010. PMID 15383274.
- ↑ Folta-Stogniew, E; O'Malley, S; Gupta, R; Anderson, KS; Radding, CM (Sep 24, 2004). "Exchange of DNA base pairs that coincides with recognition of homology promoted by E. coli RecA protein.". Molecular Cell 15 (6): 965–75. doi:10.1016/j.molcel.2004.08.017. PMID 15383285.
- ↑ Bischerour, J.; Lu, C.; Roth, D. B.; Chalmers, R. (31 August 2009). "Base Flipping in V(D)J Recombination: Insights into the Mechanism of Hairpin Formation, the 12/23 Rule, and the Coordination of Double-Strand Breaks". Molecular and Cellular Biology 29 (21): 5889–5899. doi:10.1128/MCB.00187-09. PMID 19720743.
- ↑ Klose, Robert J.; Adrian P. Bird (2006). "Genomic DNA methylation: the mark and its mediators". Trends in Biochemical Sciences 31 (2): 89–97. doi:10.1016/j.tibs.2005.12.008. ISSN 0968-0004. PMID 16403636.
- ↑ Nakao, M (2001). "Epigenetics: Interaction of DNA Methylation and Chromatin". Gene 278 (1–2): 25–31. doi:10.1016/s0378-1119(01)00721-1. PMID 11707319.
- ↑ Plass, C; Soloway, PD (2002). "DNA Methylation, Imprinting and Cancer". Eur J Hum Genet 10 (1): 6–16. doi:10.1038/sj.ejhg.5200768. PMID 11896451.
- ↑ Esteller, M; Herman, JG (2002). "Cancer as an Epigenetic Disease: DNA Methylation and Chromatin Alterations in Human Tumours". J Pathol 196 (1): 1–7. doi:10.1002/path.1024. PMID 11748635.
- ↑ "Biology and Activity of Restriction Endonucleases". http://arbl.cvmbs.colostate.edu/hbooks/genetics/biotech/enzymes/renzymes.html.
- ↑ Horton, John R.; Zhang, Xing; Maunus, Robert; Yang, Zhe; Wilson, Geoffrey; Roberts, Richard; Cheng, Xiaodong (2006). "DNA Nicking by HinP1I Endonuclease: Bending, Base Flipping and Minor Groove Expansion". Nucleic Acids Research 34 (3): 939–948. doi:10.1093/nar/gkj484. PMID 16473850.
- ↑ X-ray crystallography
- ↑ Kumar, S; Cheng, X; Pflugrath, JW; Roberts, RJ (1992). "Purification, Crystallization, and Preliminary X-ray Diffraction Analysis of an M.HhaI-AdoMet Complex". Biochemistry 31 (36): 8648–8653. doi:10.1021/bi00151a035. PMID 1390649.
- ↑ 32.0 32.1 Cheng, X; Kumar, S; Posfai, J; Pflugrath, JW; Roberts, RJ (1993). "Crystal Structure of the HhaI DNA Methyltransferase Complexed with S-adenosyl-L-methionine". Cell 74 (2): 299–307. doi:10.1016/0092-8674(93)90421-l. PMID 8343957.
- ↑ Brunger A.T. (1992)"X-PLOR, Version 3.1 : A system for x-ray crystallography and NMR"(New Haven, Connecticut: Yale University Press)
- ↑ Fitzgerald, P.M.D. (1988). "MERLOT, an integral package of computer programs for the determination of crystal structure by molecular replacement". J. Appl. Crystallogr. 21 (3): 273–288. doi:10.1107/s0021889887012299.
- ↑ NMR spectroscopy
- ↑ Gueron, M., and J. L. Leroy. 1995. Studies of basepair kinetics by NMR measurement of proton exchange. In Nuclear Magnetic Resonance And Nucleic Acids. Academic Press, San Diego, CA.
- ↑ Leijon, M.; Graslund, A. (1992). "Effects of sequence and length on imino proton-exchange and basepair opening kinetics in DNA oligonucleotide duplexes". Nucleic Acids Res 20 (20): 5339–5343. doi:10.1093/nar/20.20.5339. PMID 1331987.
- ↑ Klimasaukas, Salius, and Zita Liutkeviciute. "Experimental Approaches to Study DNA Base Flipping." DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Landes Bioscience, 2009. 37-50. Web. 16 Mar. 2014. <https://www.landesbioscience.com/pdf/04GrosjeanKlimasauskas.pdf >.
- ↑ Grosjean, [edited by] Henri (2009). DNA and RNA modification enzymes : structure, mechanism, function and evolution. Austin, Tex.: Landes Bioscience. p. 43. ISBN 978-1-58706-329-9. http://www.landesbioscience.com/pdf/04GrosjeanKlimasauskas.pdf.
- ↑ Holz, B (15 February 1998). "2-Aminopurine as a fluorescent probe for DNA base flipping by methyltransferases". Nucleic Acids Research 26 (4): 1076–1083. doi:10.1093/nar/26.4.1076. PMID 9461471.
- ↑ Yang, K; Matsika, S; Stanley, RJ (Sep 6, 2007). "6MAP, a fluorescent adenine analogue, is a probe of base flipping by DNA photolyase.". The Journal of Physical Chemistry B 111 (35): 10615–25. doi:10.1021/jp071035p. PMID 17696385.
- ↑ Yang, K; Stanley, RJ (May–Jun 2008). "The extent of DNA deformation in DNA photolyase-substrate complexes: a solution state fluorescence study.". Photochemistry and Photobiology 84 (3): 741–9. doi:10.1111/j.1751-1097.2007.00251.x. PMID 18086248.
- ↑ Berry, David A.; Jung, Kee-Yong; Wise, Dean S.; Sercel, Anthony D.; Pearson, William H.; Mackie, Hugh; Randolph, John B.; Somers, Robert L. (March 2004). "Pyrrolo-dC and pyrrolo-C: fluorescent analogs of cytidine and 2′-deoxycytidine for the study of oligonucleotides". Tetrahedron Letters 45 (11): 2457–2461. doi:10.1016/j.tetlet.2004.01.108.
- ↑ Neely, R. K.; Tamulaitis, G.; Chen, K.; Kubala, M.; Siksnys, V.; Jones, A. C. (8 September 2009). "Time-resolved fluorescence studies of nucleotide flipping by restriction enzymes". Nucleic Acids Research 37 (20): 6859–6870. doi:10.1093/nar/gkp688. PMID 19740769.
- ↑ Serva, S (1 August 1998). "Chemical display of thymine residues flipped out by DNA methyltransferases". Nucleic Acids Research 26 (15): 3473–3479. doi:10.1093/nar/26.15.3473. PMID 9671807.
- ↑ Daujotyte, D.; Liutkeviciute, Z.; Tamulaitis, G.; Klimasauskas, S. (15 April 2008). "Chemical mapping of cytosines enzymatically flipped out of the DNA helix". Nucleic Acids Research 36 (10): e57. doi:10.1093/nar/gkn200. PMID 18450817.
 |