Biology:Uracil-DNA glycosylase
 Generic protein structure example |
Uracil-DNA glycosylase (also known as UNG or UDG) is an enzyme. Its most important function is to prevent mutagenesis by eliminating uracil from DNA molecules by cleaving the N-glycosidic bond and initiating the base-excision repair (BER) pathway.
Function
The human gene encodes one of several uracil-DNA glycosylases. Alternative promoter usage and splicing of this gene leads to two different isoforms: the mitochondrial UNG1 and the nuclear UNG2.[1] One important function of uracil-DNA glycosylases is to prevent mutagenesis by eliminating uracil from DNA molecules by cleaving the N-glycosidic bond and initiating the base-excision repair (BER) pathway. Uracil bases occur from cytosine deamination or misincorporation of dUMP residues. After a mutation occurs, the mutagenic threat of uracil propagates through any subsequent DNA replication steps.[2] Once unzipped, mismatched guanine and uracil pairs are separated, and DNA polymerase inserts complementary bases to form a guanine-cytosine (GC) pair in one daughter strand and an adenine-uracil (AU) pair in the other.[3] Half of all progeny DNA derived from the mutated template inherit a shift from GC to AU at the mutation site.[3] UDG excises uracil in both AU and GU pairs to prevent propagation of the base mismatch to downstream transcription and translation processes.[3] With high efficiency and specificity, this glycosylase repairs 100–500 bases damaged daily in the human cell.[4] Human cells express five to six types of DNA glycosylases, all of which share a common mechanism of base eversion and excision as a means of DNA repair.[5]
Structure
UDG is made of a four-stranded parallel β-sheet surrounded by eight α-helices.[6] The active site comprises five highly conserved motifs that collectively catalyze glycosidic bond cleavage:[7][8]
- Water-activating loop: 63-QDPYH-67[8]
- Pro-rich loop: 165-PPPPS-169[6]
- Uracil-binding motif: 199-GVLLLN-204[6][7]
- Gly-Ser loop: 246-GS-247[6]
- Minor groove intercalation loop: 268-HPSPLS-273[6][7]
Mechanism
Glycosidic bond cleavage follows a “pinch-push-pull” mechanism using the five conserved motifs.[6]
Pinch: UDG scans DNA for uracil by nonspecifically binding to the strand and creating a kink in the backbone, thereby positioning the selected base for detection. The Pro-rich and Gly-Ser loops form polar contacts with the 3’ and 5’ phosphates flanking the examined base.[7] This compression of the DNA backbone, or “pinch,” allows for close contact between UDG and base of interest.[6]
Push: To fully assess the nucleotide identity, the intercalation loop penetrates, or pushes into, the DNA minor groove and induces a conformational change to flip the nucleotide out of the helix.[9] Backbone compression favors eversion of the now extrahelical nucleotide, which is positioned for recognition by the uracil-binding motif.[6] The coupling of intercalation and eversion helps compensate for the disruption of favorable base stacking interactions within the DNA helix. Leu272 fills the void left by the flipped nucleotide to create dispersion interactions with neighboring bases and restore stacking stability.[7]
Pull: Now accessible to the active site, the nucleotide interacts with the uracil binding motif. The active site shape complements the everted uracil structure, allowing for high substrate specificity. Purines are too large to fit in the active site, while unfavorable interactions with other pyrimidines discourage binding alternative substrates.[5] The side chain of Tyr147 interferes sterically with the thymine C5 methyl group, while a specific hydrogen bond between the uracil O2 carbonyl and Gln144 discriminates against a cytosine substrate, which lacks the necessary carbonyl.[5] Once uracil is recognized, cleavage of the glycosidic bond proceeds according to the mechanism below.
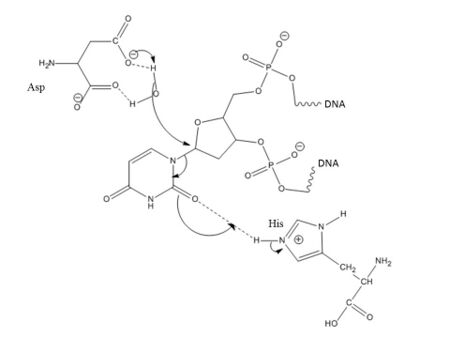
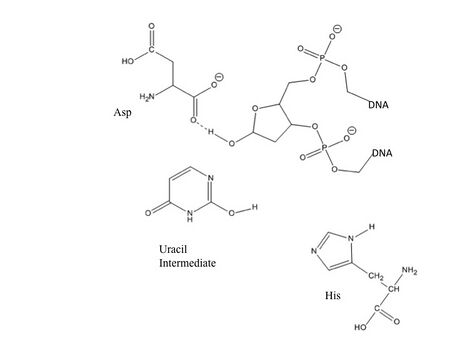
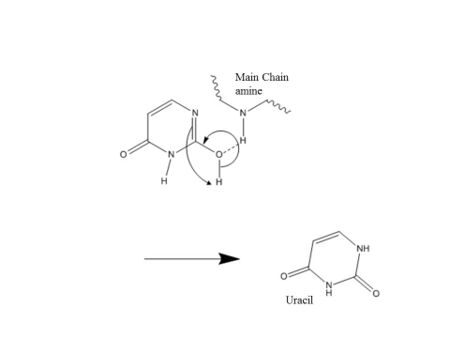
The position of the residues that activate the water nucleophile and protonate the uracil leaving group are widely debated, though the most commonly followed mechanism employs the water activating loop detailed in the enzyme structure.[8][10] Regardless of position, the identities of the aspartic acid and histidine residues are consistent across catalytic studies.[6][7][8][10][11]
Laboratory use
Uracil N-glycosylase (UNG) is utilized to eliminate carryover polymerase chain reaction (PCR) products in PCR. This method modifies PCR products such that in a new reaction, any residual products from previous PCR amplifications will be digested and prevented from amplifying, but the true DNA templates will be unaffected.[12] PCR synthesizes abundant amplification products each round, but contamination of further rounds of PCR with trace amounts of these products, called carry-over contamination, yields false positive results. Carry-over contamination from some previous PCR can be a significant problem, due both to the abundance of PCR products, and to the ideal structure of the contaminant material for re-amplification. However carry-over contamination can be controlled by the following two steps: (i) incorporating dUTP in all PCR products (by substituting dUTP for dTTP, or by incorporating uracil during synthesis of primers; and (ii) treating all subsequent fully preassembled starting reactions with uracil DNA glycosylase (UDG), followed by thermal inactivation of UDG. UDG cleaves the uracil base from the phosphodiester backbone of uracil-containing DNA, but has no effect on natural (i.e., thymine-containing) DNA. The resulting apyrimidinic sites block replication by DNA polymerases, and are very labile to acid/base hydrolysis. Because UDG does not react with dTTP, and is also inactivated by heat denaturation prior to the actual PCR, carry-over contamination of PCRs can be controlled effectively if the contaminants contain uracils in place of thymines.[2]
Uracil N-glycosylase was also used in a study to detect evidence of ongoing low-level metabolic activity and DNA repair in ancient bacteria.[13] Long-term survival of bacteria can occur either through endospore formation (in which the bacterium enters total dormancy, with no metabolic activity at all taking place, and, thus, no DNA repair), or else through reduction of metabolic activity to a very low rate, just sufficient to carry out ongoing DNA repair and prevent the depletion of other unstable molecules (such as ATP), in which the microbe is able to repair damage to its DNA but also continues to slowly consume nutrients.[13] DNA sequences from bacteria in permafrost were amplified using PCR. One series of runs amplified the DNA sequences as-is (to detect all live bacterial DNA in the samples), while the other series looked specifically for DNA that had been undergoing ongoing repair; to do this, the DNA was treated with UNG to remove uracils. This prevented amplification of unrepaired DNA in two ways: firstly, the abasic sites generated by the removal of uracils prevented the DNA polymerase used in PCR from proceeding past the site of damage, while these abasic sites also directly weakened the DNA and made it more likely to fragment upon heating.[13] In this way, the researchers were able to show evidence of ongoing DNA repair in high-GC Gram-positive bacteria up to 600,000 years old.[13]
Uracil N glycosylase has also been used in a method for cloning of PCR amplified DNA fragments. In this method the primers used in PCR are synthesized with uracil residues instead of thymine. When these primers are incorporated into PCR amplified fragments the primer sequence becomes susceptible to digestion with Uracil N Glycosylase and produce 3' protruding ends that can be annealed to an appropriately prepared vector DNA. The resulting chimeric molecules can be transformed into competent cells with high efficiency, without the need for in vitro ligation.[14]
Interactions
Uracil-DNA glycosylase has been shown to interact with RPA2.[15]
References
- ↑ "Entrez Gene: UNG uracil-DNA glycosylase". https://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=7374.
- ↑ 2.0 2.1 "Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions". Gene 93 (1): 125–8. Sep 1990. doi:10.1016/0378-1119(90)90145-H. PMID 2227421.
- ↑ 3.0 3.1 3.2 "Structure and function in the uracil-DNA glycosylase superfamily". Mutation Research 460 (3–4): 165–81. Aug 2000. doi:10.1016/S0921-8777(00)00025-2. PMID 10946227.
- ↑ "A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA". Nature 384 (6604): 87–92. Nov 1996. doi:10.1038/384087a0. PMID 8900285. Bibcode: 1996Natur.384...87S.
- ↑ 5.0 5.1 5.2 "Suppression of spontaneous mutagenesis in human cells by DNA base excision-repair". Mutation Research 462 (2–3): 129–35. Apr 2000. doi:10.1016/S1383-5742(00)00024-7. PMID 10767624.
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 6.6 6.7 6.8 "Lessons learned from structural results on uracil-DNA glycosylase". Mutation Research 460 (3–4): 183–99. Aug 2000. doi:10.1016/S0921-8777(00)00026-4. PMID 10946228.
- ↑ 7.0 7.1 7.2 7.3 7.4 7.5 "Uracil-DNA glycosylase: Structural, thermodynamic and kinetic aspects of lesion search and recognition". Mutation Research 685 (1–2): 11–20. Mar 2010. doi:10.1016/j.mrfmmm.2009.10.017. PMID 19909758.
- ↑ 8.0 8.1 8.2 8.3 "Complexes of the uracil-DNA glycosylase inhibitor protein, Ugi, with Mycobacterium smegmatis and Mycobacterium tuberculosis uracil-DNA glycosylases". Microbiology 149 (Pt 7): 1647–58. Jul 2003. doi:10.1099/mic.0.26228-0. PMID 12855717.
- ↑ "Crystal structure and mutational analysis of human uracil-DNA glycosylase: structural basis for specificity and catalysis". Cell 80 (6): 869–78. Mar 1995. doi:10.1016/0092-8674(95)90290-2. PMID 7697717.
- ↑ 10.0 10.1 "Crystal structure of vaccinia virus uracil-DNA glycosylase reveals dimeric assembly". BMC Structural Biology 7: 45. 2007. doi:10.1186/1472-6807-7-45. PMID 17605817.
- ↑ "The structural basis of specific base-excision repair by uracil-DNA glycosylase". Nature 373 (6514): 487–93. Feb 1995. doi:10.1038/373487a0. PMID 7845459. Bibcode: 1995Natur.373..487S.
- ↑ "Support Centers - Thermo Fisher Scientific". https://abcommunity.thermofisher.com/community/real-time_pcr/blog/2013/08/02/what-is-ung-and-how-does-it-work.
- ↑ 13.0 13.1 13.2 13.3 Johnson SS; Hebsgaard MB; Christensen TR; Mastepanov M; Nielsen R; Munch K; Brand T; Gilbert MT et al. (September 2007). "Ancient bacteria show evidence of DNA repair". PNAS 104 (36): 14401–5. doi:10.1073/pnas.0706787104. PMID 17728401. Bibcode: 2007PNAS..10414401J.
- ↑ .Analytical biochemistry 1992 ;206(1):91-7.
- ↑ "A sequence in the N-terminal region of human uracil-DNA glycosylase with homology to XPA interacts with the C-terminal part of the 34-kDa subunit of replication protein A". The Journal of Biological Chemistry 272 (10): 6561–6. Mar 1997. doi:10.1074/jbc.272.10.6561. PMID 9045683.
 |