Biology:Rozellida
| Rozellida | |
|---|---|
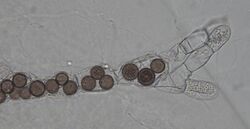
| |
| Rozella allomycis parasitizing the chytrid Allomyces sp. | |
| Scientific classification | |
| Domain: | Eukaryota
|
| (unranked): | Opisthokonta
|
| (unranked): | |
| Kingdom: | |
| Phylum: | Rozellomycota
|
| Class: | Rozellidea
|
| Order: | Rozellida Cavalier-Smith 2013
|
| Genus | |
| |
| Synonyms | |
| |
Cryptomycota ('hidden fungi'), Rozellida, or Rozellomycota are a clade of micro-organisms that are either fungi or a sister group to fungi. They differ from classical fungi in that they lack chitinous cell walls at any trophic stage in their lifecycle, as reported by Jones and colleagues in 2011.[1][2] Despite their unconventional feeding habits,[clarification needed] chitin has been observed in the inner layer of resting spores, and in immature resting spores for some species of Rozella, as indicated with calcofluor-white stain as well as the presence of a fungal-specific chitin synthase gene.[3]
Rozellida were first detected as DNA sequences retrieved from a freshwater laboratory enclosure. Phylogenetic analysis of these sequences formed a unique terminal clade of then unknown affiliation provisionally called after the first clone in the clade: LKM11.[4]
The only formally described genus in the clade is Rozella, which was previously considered a chytrid. The existence of related organisms was known from environmental DNA sequences.[5]
Additional members of the group were isolated in 2011 by a team led by Thomas Richards, from the Natural History Museum in London, and also an evolutionary geneticist at the University of Exeter, UK. The team used DNA techniques to disclose the existence of unknown genetic material dredged from the university pond. Once they had a few unknown sequences they fluorescently labeled small DNA sequences and let them bind to the matching DNA in the whole sample (fluorescence in situ hybridization). Under fluorescence microscopy, they could see that the possessor cells were ovoid in shape and 3–5 micrometres across. They then established that the cryptomycota were present in other samples taken from further freshwater environments, soils and marine sediments.[6][7]
The common characteristic of the clade members is that they lack the chitinous cell walls present in almost all previously discovered fungi (including microsporidia) and which are a major feature of the kingdom. Without the chitin the cryptomycota can be phagotrophic parasites that feed by attaching to, engulfing, or living inside other cells. Most known fungi feed by osmotrophy—taking in nutrients from outside the cell.[6]
Taxonomy
| Phylogeny of Rozellomycota[8] | |||||||||||||||||||||||||||||||||||||||||||||
|
- Phylum Rozellomycota [9]
- Class Rozellidea Cavalier-Smith 2013 s.s.
- Order Rozellida Cavalier-Smith 2013 s.s.
- Genus Mitosporidium Haag et al. 2014
- Genus Morellospora Corsaro et al. 2020
- Genus Nucleophaga Dangeard 1895
- Genus Paramicrosporidium Corsaro et al. 2014
- Genus Rozella Cornu 1872
- Order Rozellida Cavalier-Smith 2013 s.s.
- Class Rozellidea Cavalier-Smith 2013 s.s.
References
- ↑ "Validation and justification of the phylum name Cryptomycota phyl. nov.". IMA Fungus 2 (2): 173–7. 2011. doi:10.5598/imafungus.2011.02.02.08. PMID 22679602.
- ↑ "Discovery of novel intermediate forms redefines the fungal tree of life". Nature 474 (7350): 200–3. 2011. doi:10.1038/nature09984. PMID 21562490.
- ↑ "No jacket required—New fungal lineage defies dress code". BioEssays 34 (2): 94–102. 2011. doi:10.1002/bies.201100110. PMID 22131166.
- ↑ "Detritus-dependent development of the microbial community in an experimental system: Qualitative analysis by denaturing gradient gel electrophoresis". Applied and Environmental Microbiology 65 (6): 2478–84. 1999. doi:10.1128/AEM.65.6.2478-2484.1999. PMID 10347030. Bibcode: 1999ApEnM..65.2478V.
- ↑ "The environmental clade LKM11 and Rozella form the deepest branching clade of fungi". Protist 161 (1): 116–21. 2010. doi:10.1016/j.protis.2009.06.005. PMID 19674933. http://doc.rero.ch/record/17571/files/Lara_Enrique_-_The_Environmental_Clade_LKM11_and_Rozella_20100319.pdf.
- ↑ 6.0 6.1 Turner M. (11 May 2011). "The evolutionary tree of fungi grows a new branch". Nature News. doi:10.1038/news.2011.285.
- ↑ Ghosh P. (11 May 2011). "'Missing link' fungi found in Devon pond". BBC News. https://www.bbc.co.uk/news/mobile/science-environment-13363932.
- ↑ "Outline of Fungi and fungus-like taxa". Mycosphere 11 (1): 1060–1456. 2020. doi:10.5943/mycosphere/11/1/8. ISSN 2077-7019. http://www.mycosphere.org/pdf/MYCOSPHERE_11_1_8-1.pdf.
- ↑ Karpov (2014). "Morphology, phylogeny and ecology of the aphelids (Aphelidea, Opisthokonta) and proposal for the new superphylum Opisthosporidia". Frontiers in Microbiology 5: 112. doi:10.3389/fmicb.2014.00112. PMID 24734027.
Wikidata ☰ Q1142300 entry
 |

