Biology:Phi X 174
| Escherichia virus ΦX174 | |
|---|---|
| |
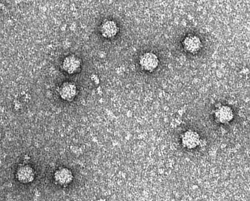
| Electron micrograph of phage ΦX174 | |
| Virus classification Error creating thumbnail: Unable to save thumbnail to destination
| |
| (unranked): | Virus |
| Realm: | Monodnaviria |
| Kingdom: | Sangervirae |
| Phylum: | Phixviricota |
| Class: | Malgrandaviricetes |
| Order: | Petitvirales |
| Family: | Microviridae |
| Genus: | Sinsheimervirus |
| Species: | Escherichia virus ΦX174
|
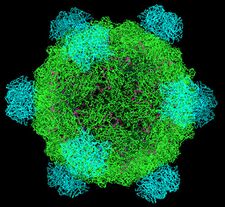
The phi X 174 (or ΦX174) bacteriophage is a single-stranded DNA (ssDNA) virus that infects Escherichia coli. This virus was isolated in 1935 by Nicolas Bulgakov [1] in Félix d'Hérelle's laboratory at the Pasteur Institute, from samples collected in Paris sewers. Its characterization and the study of its replication mechanism were carried out from the 1950s onwards. It was the first DNA-based genome to be sequenced. This work was completed by Fred Sanger and his team in 1977.[2] In 1962, Walter Fiers and Robert Sinsheimer had already demonstrated the physical, covalently closed circularity of ΦX174 DNA.[3] Nobel prize winner Arthur Kornberg used ΦX174 as a model to first prove that DNA synthesized in a test tube by purified enzymes could produce all the features of a natural virus, ushering in the age of synthetic biology.[4][5] In 1972–1974, Jerard Hurwitz, Sue Wickner, and Reed Wickner with collaborators identified the genes required to produce the enzymes to catalyze conversion of the single stranded form of the virus to the double stranded replicative form.[6] In 2003, it was reported by Craig Venter's group that the genome of ΦX174 was the first to be completely assembled in vitro from synthesized oligonucleotides.[7] The ΦX174 virus particle has also been successfully assembled in vitro.[8] In 2012, it was shown how its highly overlapping genome can be fully decompressed and still remain functional.[9]
Genome
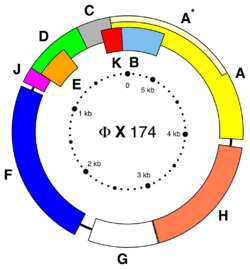
This bacteriophage has a [+] sense circular single-stranded DNA genome of 5,386 nucleotides.[10] The genome GC-content is 44% and 95% of nucleotides belong to coding genes. Because of the balance base pattern of the genome, it is used as the control DNA for Illumina sequencers.[citation needed]
Genes
ΦX174 encodes 11 genes, named as consecutive letters of the alphabet in the order they were discovered, with the exception of A* which is an alternative start codon within the large A genes. Only genes A* and K are thought to be non-essential, although there is some doubt about A* because its start codon could be changed to ATT but not any other sequence.[11] It is now known that the ATT is still likely capable of producing protein[12] within E. coli and therefore this gene may in fact be essential.
The first half of the ΦX174 genome features high levels of gene overlap[13] with eight out of 11 genes overlapping by at least one nucleotide.[2] These overlaps have been shown to be non-essential [9] although the refactored phage with all gene overlaps removed had decreased fitness from wild-type.[14]
Phage ΦX174 has been used to try to establish the absence of undiscovered genetic information through a "proof by synthesis" approach.[15]
Transcriptome
In 2020, the transcriptome of ΦX174 was generated.[16] Notable features of the ΦX174 transcriptome is a series of up to four relatively weak promoters in series with up to four Rho-independent (intrinsic) terminators and one Rho-dependent terminator.[citation needed]
Proteins
ΦX174 encodes 11 proteins.
| Protein | Copies | Function[17] |
|---|---|---|
| A | — | Nicks RF DNA to initiate rolling circle replication; ligates ends of linear phage DNA to form single-stranded circular DNA |
| A* | — | Inhibits host cell DNA replication; blocks superinfecting phage; not essential |
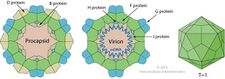
| B | 60 in procapsid | Internal scaffolding protein involved in procapsid assembly |
| C | — | DNA packaging |
| D | 240 in procapsid | External scaffolding protein involved in procapsid assembly |
| E | — | Host cell lysis |
| F | 60 in virion | Major capsid protein |
| G | 60 in virion | Major spike protein |
| H | 12 in virion | DNA pilot protein (or minor spike protein) |
| J | 60 in virion | Binds to new single-stranded phage DNA; accompanies phage DNA into procapsid |
| K | — | Optimizes burst size; not essential |
Proteome
Identification of all ΦX174 proteins using mass spectrometry has recently been reported.[14]
Infection Cycle
Infection begins when G protein binds to lipopolysaccharides on the bacterial host cell surface. H protein (or the DNA Pilot Protein) pilots the viral genome through the bacterial membrane of E.coli bacteria[18] most likely via a predicted N-terminal transmembrane domain helix.[19] However, it has become apparent that H protein is a multifunctional protein.[20] This is the only viral capsid protein of ΦX174 to lack a crystal structure for a couple of reasons. It has low aromatic content and high glycine content, making the protein structure very flexible and in addition, individual hydrogen atoms (the R group for glycines) are difficult to detect in protein crystallography. Additionally, H protein induces lysis of the bacterial host at high concentrations as the predicted N-terminal transmembrane helix easily pokes holes through the bacterial wall. By bioinformatics, this protein contains four predicted coiled-coil domains which has a significant homology to known transcription factors. Additionally, it was determined that de novo H protein was required for optimal synthesis of other viral proteins.[21] Mutations in H protein that prevent viral incorporation, can be overcome when excess amounts of protein B, the internal scaffolding protein, are supplied.[citation needed]
The DNA is ejected through a hydrophilic channel at the 5-fold vertex.[22] It is understood that H protein resides in this area but experimental evidence has not verified its exact location. Once inside the host bacterium, replication of the [+] ssDNA genome proceeds via negative sense DNA intermediate. This is done as the phage genome supercoils and the secondary structure formed by such supercoiling attracts a primosome protein complex. This translocates once around the genome and synthesizes a [−]ssDNA from the positive original genome. [+]ssDNA genomes to package into viruses are created from this by a rolling circle mechanism. This is the mechanism by which the double stranded supercoiled genome is nicked on the positive strand by a virus-encoded A protein, also attracting a bacterial DNA polymerase (DNAP) to the site of cleavage. DNAP uses the negative strand as a template to make positive sense DNA. As it translocates around the genome it displaces the outer strand of already-synthesised DNA, which is immediately coated by SSBP proteins. The A protein cleaves the complete genome every time it recognises the origin sequence.[citation needed]
As D protein is the most abundant gene transcript, it is the most protein in the viral procapsid. Similarly, gene transcripts for F, J, and G are more abundant than for H as the stoichiometry for these structural proteins is 5:5:5:1. The primosomes are protein complexes which attach/bind the enzyme helicase on the template. Primosomes gives RNA primers for DNA synthesis to strands.[citation needed]
Phylogenetics and diversity
ΦX174 is closely related to other microviridae, especially the NC phage (e.g. NC1, NC7, NC11, NC16, NC37, NC5, NC41, NC56, NC51, etc.) and more distantly related to the G4-like phages and even more distantly related to the α3-like phage. Rokyta et al. 2006 presented a phylogenetic tree of their relationships.[23]
Uses
Experimental evolution
ΦX174 has been used as a model organism in many evolution experiments.[24]
Biotechnology
ΦX174 is regularly used as a positive control in DNA sequencing due to its relatively small genome size in comparison to other organisms, its relatively balanced nucleotide content — about 23% G, 22% C, 24% A, and 31% T, i.e., 45% G+C and 55% A+T, see the accession NC_001422.1[10] for its 5,386 nucleotide long sequence. Illumina's sequencing instruments use ΦX174 as a positive control,[25] and a single Illumina sequencing run can cover the ΦX174 genome several million times over, making this very likely the most heavily sequenced genome in history.[citation needed]
ΦX174 is also used to test the resistance of personal protective equipment to bloodborne viruses.[26]
ΦX174 has also been modified to enable peptide display (phage display) from the viral capsid G protein.[27]
Synthetic Biology
The ΦX174 genome was the first phage to be cloned in yeast,[9] which provides a convenient drydock for genome modifications.[28] ΦX174 was also the first genome to be fully decompressed, having all gene overlaps removed.[13] The effect of these changes resulted in significantly reduced host attachment, protein expression dysregulation, and heat sensitivity.[14]
See also
References
- ↑ Lacković, Zdravko; Toljan, Karlo (2020-12-20). "Vladimir Sertić: forgotten pioneer of virology and bacteriophage therapy" (in en). Notes and Records: the Royal Society Journal of the History of Science 74 (4): 567–578. doi:10.1098/rsnr.2019.0010. ISSN 0035-9149. PMID 33177747. PMC 7653334. https://royalsocietypublishing.org/doi/10.1098/rsnr.2019.0010.
- ↑ 2.0 2.1 "Nucleotide sequence of bacteriophage phi X174 DNA". Nature 265 (5596): 687–95. February 1977. doi:10.1038/265687a0. PMID 870828. Bibcode: 1977Natur.265..687S.
- ↑ "The structure of the DNA of bacteriophage phi-X174. III. Ultracentrifugal evidence for a ring structure". Journal of Molecular Biology 5 (4): 424–34. October 1962. doi:10.1016/S0022-2836(62)80031-X. PMID 13945085.
- ↑ National Library of Medicine Profiles in Science. The Arthur Kornberg Papers. "Creating Life in the Test Tube," 1959-1970. link[non-primary source needed]
- ↑ "Enzymatic synthesis of DNA, XXIV. Synthesis of infectious phage phi-X174 DNA". Proceedings of the National Academy of Sciences of the United States of America 58 (6): 2321–8. December 1967. doi:10.1073/pnas.58.6.2321. PMID 4873588. Bibcode: 1967PNAS...58.2321G.
- ↑ "Conversion of phiX174 viral DNA to double-stranded form by purified Escherichia coli proteins". Proceedings of the National Academy of Sciences of the United States of America 71 (10): 4120–4. October 1974. doi:10.1073/pnas.71.10.4120. PMID 4610569.
- ↑ "Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides". Proceedings of the National Academy of Sciences of the United States of America 100 (26): 15440–5. December 2003. doi:10.1073/pnas.2237126100. PMID 14657399. Bibcode: 2003PNAS..10015440S.
- ↑ "In VITRO ASSEMBLY of the øX174 procapsid from external scaffolding protein oligomers and early pentameric assembly intermediates". Journal of Molecular Biology 412 (3): 387–96. September 2011. doi:10.1016/j.jmb.2011.07.070. PMID 21840317.
- ↑ 9.0 9.1 9.2 "A fully decompressed synthetic bacteriophage øX174 genome assembled and archived in yeast". Virology 434 (2): 278–84. December 2012. doi:10.1016/j.virol.2012.09.020. PMID 23079106.
- ↑ 10.0 10.1 10.2 Enterobacteria phage phiX174 sensu lato, complete genome. "Complete genome: accession NC_001422", National Center for Biotechnology Information. Retrieved on 30 January 2016.
- ↑ "Alteration of the ATG start codon of the A protein of bacteriophage phi X174 into an ATT codon yields a viable phage indicating that A protein is not essential for phi X174 reproduction". FEBS Letters 218 (1): 119–25. June 1987. doi:10.1016/0014-5793(87)81030-x. PMID 2954853.
- ↑ "Measurements of translation initiation from all 64 codons in E. coli". Nucleic Acids Research 45 (7): 3615–3626. April 2017. doi:10.1093/nar/gkx070. PMID 28334756.
- ↑ 13.0 13.1 Wright, Bradley W.; Molloy, Mark P.; Jaschke, Paul R. (2021-10-05). "Overlapping genes in natural and engineered genomes" (in en). Nature Reviews Genetics 23 (3): 154–168. doi:10.1038/s41576-021-00417-w. ISSN 1471-0064. PMID 34611352.
- ↑ 14.0 14.1 14.2 "Genome Modularization Reveals Overlapped Gene Topology Is Necessary for Efficient Viral Reproduction". ACS Synthetic Biology 9 (11): 3079–3090. November 2020. doi:10.1021/acssynbio.0c00323. PMID 33044064.
- ↑ "Definitive demonstration by synthesis of genome annotation completeness". Proceedings of the National Academy of Sciences of the United States of America 116 (48): 24206–24213. November 2019. doi:10.1073/pnas.1905990116. PMID 31719208.
- ↑ "A high-resolution map of bacteriophage ϕX174 transcription". Virology 547: 47–56. August 2020. doi:10.1016/j.virol.2020.05.008. PMID 32560904.
- ↑ Fane, Bentley A.; Brentlinger, Karie L.; Burch, April D.; Chen, Min; Hafenstein, Susan; Moore, Erica; Novak, Christopher R.; Uchiyama, Asako (2006). "ɸX174 et al., the Microviridae". in Calender, Richard. The Bacteriophages (2nd ed.). New York: Oxford Univ. Press. p. 130. ISBN 978-0195148503.
- ↑ "The lipopolysaccharide receptor for bacteriophage phiX174 and S13". Virology 66 (1): 268–82. July 1975. doi:10.1016/0042-6822(75)90197-x. PMID 1094681.
- ↑ "The HMMTOP transmembrane topology prediction server". Bioinformatics 17 (9): 849–50. September 2001. doi:10.1093/bioinformatics/17.9.849. PMID 11590105.
- ↑ "Uncoupling the functions of a multifunctional protein: the isolation of a DNA pilot protein mutant that affects particle morphogenesis". Virology 411 (1): 9–14. March 2011. doi:10.1016/j.virol.2010.12.026. PMID 21227478.
- ↑ "The expression of N-terminal deletion DNA pilot proteins inhibits the early stages of phiX174 replication". Journal of Virology 83 (19): 9952–6. October 2009. doi:10.1128/JVI.01077-09. PMID 19640994.
- ↑ "Atomic structure of single-stranded DNA bacteriophage phi X174 and its functional implications". Nature 355 (6356): 137–43. January 1992. doi:10.1038/355137a0. PMID 1370343. Bibcode: 1992Natur.355..137M.
- ↑ "Horizontal gene transfer and the evolution of microvirid coliphage genomes". Journal of Bacteriology 188 (3): 1134–42. February 2006. doi:10.1128/JB.188.3.1134-1142.2006. PMID 16428417.
- ↑ "Experimental evolution of viruses: Microviridae as a model system". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 365 (1552): 2495–501. August 2010. doi:10.1098/rstb.2010.0053. PMID 20643739.
- ↑ "Using a PhiX Control for HiSeq® Sequencing Runs". Illumina. https://support.illumina.com/sequencing/sequencing_kits/phix_control_v3.html.
- ↑ "PPE-Info – Standard Details". https://wwwn.cdc.gov/PPEInfo/Standards/Info/ASTMF181907(2013).
- ↑ "PhiXing-it, displaying foreign peptides on bacteriophage ΦX174". Virology 488: 242–8. January 2016. doi:10.1016/j.virol.2015.11.021. PMID 26655242.
- ↑ "Engineering Modular Viral Scaffolds for Targeted Bacterial Population Editing". Cell Systems 1 (3): 187–196. September 2015. doi:10.1016/j.cels.2015.08.013. PMID 26973885.
External links
- "Bacteriophage phiX174". Molecule of the Month. RCSB-PDB. February 2000. http://www.rcsb.org/pdb/101/motm.do?momID=2.
Wikidata ☰ Q1063448 entry
 |